Digital PCR (dPCR) enables precise, highly sensitive quantification of nucleic acids. Traditional PCR is an end-point analysis that is semi-quantitative because the amplified product is detected by agarose gel electrophoresis. Real-time PCR (or qPCR) uses fluorescence-based detection to allow the measurement of accumulated amplified product as the reaction progresses. qPCR requires normalization to controls (either to a reference or to a standard curve), allowing only relative quantification. Furthermore, variations in amplification efficiency may affect qPCR results.
Digital PCR builds on traditional PCR amplification and fluorescent-probe–based detection methods to provide highly sensitive absolute quantification of nucleic acids without the need for standard curves. In the Droplet Digital™ PCR System, a PCR sample is partitioned into 20,000 droplets. After amplification, droplets containing target sequence are detected by fluorescence and scored as positive, and droplets without fluorescence are scored as negative. Poisson statistical analysis of the numbers of positive and negative droplets yields absolute quantitation of the target sequence.
Page Contents
The concept of digital PCR was first described in 1992 by Sykes et al., who recognized that the combination of limiting dilution, end-point PCR, and Poisson statistics could yield an absolute measure of nucleic acid concentration (Sykes et al. 1992). Subsequently, Vogelstein and Kinzler at Johns Hopkins University developed a method whereby a sample is diluted and partitioned to the extent that single template molecules can be amplified individually, each in a separate partition, and the products detected using fluorescent probes (Vogelstein and Kinzler 1999). The term “digital PCR” was coined to describe this novel method.
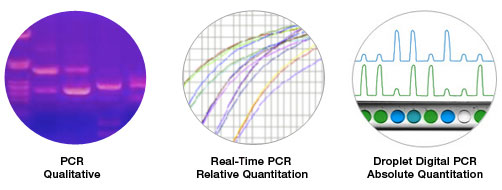
Digital PCR represents a generation of PCR that enables absolute quantification of target sequences.
Digital PCR improves upon the sensitivity of qPCR and enables the detection of rare events such as single-nucleotide mutations in a population of wild-type sequences. In conventional qPCR, the signal from wild-type sequences can dominate and obscure the signal from the rare sequence. By minimizing the effects of competition between targets, digital PCR overcomes the difficulties inherent to amplifying rare sequences, and allows for sensitive and precise absolute quantification of nucleic acids.
A critical step in digital PCR is sample partitioning — the division of each sample into discrete subunits prior to amplification by PCR. The sample is prepared in a manner similar to that for real-time PCR but is then separated into thousands of partitions, each ideally containing either zero or one (or, at most, a few) template molecules. Each partition behaves as an individual PCR reaction and, as with real-time PCR, fluorescent probes are used to identify amplified target DNA. Each partition can then be readily analyzed after amplification to determine whether or not it contains the target sequence. Samples containing amplified product are considered positive (1, fluorescent), and those without product, and thus with little or no fluorescence, are negative (0). The ratio of positives to negatives in each sample is the basis of quantification. Unlike real-time qPCR, digital PCR does not rely on the number of amplification cycles to determine the initial amount of template nucleic acid in each sample; rather, it relies on Poisson statistics to determine the absolute template quantity.
The unique sample partitioning step of digital PCR, paired with Poisson statistical data analysis, allows higher precision than traditional PCR and qPCR methods. Accordingly, digital PCR is particularly well suited for applications that require the detection of small amounts of input nucleic acid or finer resolution of target amounts among samples, for example, rare sequence detection, copy number variation (CNV) analysis, and gene expression analysis of the rare targets.
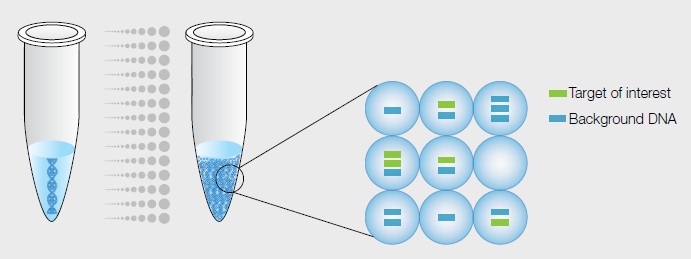
Sample partitioning is the key to digital PCR. In the Droplet Digital PCR (ddPCR™) System, a single PCR sample is partitioned into 20,000 droplets. Template molecules are distributed randomly among droplets, such that some droplets have no template molecules and others have one or more. Each droplet undergoes PCR amplification and analysis separately, and the droplets are then individually counted and scored as positive or negative for fluorescence.
The methods described by Sykes et al. (1992) and Vogelstein and Kinzler (1999) have been modified and made commercially available in several formats. Methods available for digital PCR include PCR amplification on a microfluidic chip (Warren et al. 2006, Ottesen et al. 2006, Fan and Quake 2007). Other systems involve separation onto microarrays (Morrison et al. 2006) or spinning microfluidic discs (Sundberg et al. 2010) and droplet techniques based on oil-water emulsions (Hindson et al., 2011), as in Bio-Rad’s QX200 Droplet Digital PCR System
Introduction to Digital PCR | Bio-Rad
Sample partitioning in the ddPCR System allows the sensitive, specific detection of single template molecules and precise quantification while mitigating the effects of target competition, making PCR amplification less sensitive to inhibition and greatly improving the discriminatory capacity of assays that differ by only a single nucleotide. Digital PCR offers significant benefits over other PCR methods: absolute quantification and greatly enhanced sensitivity and dynamic range. Thanks to its high sensitivity, precision, and absolute quantification, digital PCR extends the range of nucleic acid analysis beyond the reach of other methods in a number of applications:
- Liquid biopsy
- Copy number variation (CNV)
- Rare sequence detection
- Gene expression and miRNA analysis
- Single-cell analysis
- Pathogen detection
- Next-generation sequencing (NGS) library analysis
- Contaminant testing in cell and gene therapy
Choosing a Mycoplasma Detection Strategy
Visit the Digital PCR Applications section for an overview of current and emerging applications of digital PCR.
 |
Download the Droplet Digital PCR Applications Guide for in-depth information about setting up experiments for these and other applications. |
Fan HC and Quake SR (2007). Detection of aneuploidy with digital polymerase chain reaction. Anal Chem 79, 7576–7579. PMID: 17715994
Hindson BJ et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83, 8604–8610. PMID: 22035192
Morrison T et al. (2006). Nanoliter high-throughput quantitative PCR. Nucleic Acids Res 34, e123. PMID: 17000636
Ottesen EA et al. (2006). Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 314, 1464–1467. PMID: 17138901
Sundberg SO et al. (2010). Spinning disk platform for microfluidic digital polymerase chain reaction. Anal Chem 82, 1546–1550. PMID: 20085301
Sykes PJ et al. (1992). Quantitation of targets for PCR by use of limiting dilution. BioTechniques 13, 444–449. PMID: 1389177
Vogelstein B and Kinzler KW (1999). Digital PCR. Proc Nat Acd Sci USA 96, 9236–9241. PMID: 10430926
Warren L et al. (2006). Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci USA 103, 17807–17812. PMID: 17098862
Baker M (2012). Digital PCR hits its stride. Nat Methods 9, 541–544.
Huggett J and Scott D (2010). Digital polymerase chain reaction: New diagnostic opportunities. European Pharmaceutical Review, Industry Focus 2010.
Kubista M (2008). Emerging real-time PCR applications. Drug Discovery World Summer 2008, 57–66.
Kubista M and Stahlberg A (2011). DNA diagnostics gets digitized. Drug Discovery World Fall 2011, 77–82.
McCaughan P and Dear PH (2010). Single-molecule genomics. J Pathol 220, 297–306. PMID: 19927313
Pohl G and Shih IeM (2004). Principle and applications of digital PCR. Expert Mol Rev Diagn 4, 41–47. PMID: 14711348