The ProteOn™ XPR36 system uses the 6 × 6 interaction array of the ProteOn sensor chips. ProteOn sensor chips contain more than just a gold layer; they are coated with a modified alginate polymer that provides a solution-like, biomimetic environment for ligand immobilization. The general-use ProteOn sensor chips (GLC, GLM, and GLH sensor chips) are functionalized with carboxylic acid groups that react with surface-exposed amines on the ligand, tethering the ligand to the sensor chip in a random orientation. The ProteOn NLC sensor chips are coated with NeutrAvidin for immobilization of biotinylated ligands, and the HTG and HTE sensor chips feature a tris-NTA surface for immobilization of histidine-tagged proteins. The respective chips can be used to capture a ligand at a site-specific location. The ProteOn LCP sensor chips are used with the LCP capturing reagent kit for liposome capture. Although choosing a suitable ProteOn sensor chip for a particular interaction requires some research and planning, this is time well spent considering the high-quality data that can be obtained with the right sensor chip.
Related Topics: Surface Plasmon Resonance and the ProteOn XPR36 System.
Page Contents
ProteOn sensor chips are built with an alginate polymer matrix bound to a thin gold film on a sensor prism. The alginate matrix can be functionalized with several different reactive groups to facilitate different immobilization chemistries. The hydrophilic nature of the alginate layer creates a solution-like environment that prevents denaturation of the immobilized ligand and non-specific adsorption of the analyte. The increased surface area of the 3-D alginate matrix provides more attachment sites than a 2-D monolayer and results in more ligand molecules being immobilized on the sensor chip surface. In addition, the molecular weight and structure of the alginate coating can be modified to create sensor chips with different surface capacities. This results in sensitive detection of interactions with minimal surface effects on binding. The surface chemistry of the ProteOn sensor chips allows the ProteOn to detect tight binding interactions down to picomolar concentrations of analytes, or analytes as small as 95 Daltons. The combination of ProteOn sensor chips with the unique 6 x 6 interaction array allows for the interaction of up to 36 separate ligand/analyte pairs on a single chip, increasing the throughput of a single experiment.
The ProteOn sensor chips are based on innovative, patent-protected surface chemistry. The general-use ProteOn sensor chips (GLC, GLM, and GLH sensor chips) come functionalized with carboxylic acid groups to facilitate amine coupling of protein ligands via surface-exposed amine groups. In addition to serving as attachment sites, the carboxylic acid groups serve to concentrate the ligand at the surface of the sensor chip as the negatively charged carboxyl groups attract proteins rendered positively charged via incubation in an acidic buffer. Activation of carboxylic acid groups for ligand immobilization is done using carbodiimide chemistry with the reagents EDC and sulfo-NHS. Once activated, the resulting sulfo-NHS esters are highly amine-reactive and react with free amines exposed on the ligand to immobilize it to the sensor chip. Any unreacted carboxyl groups that remain activated during the immobilization step are deactivated with ethanolamine to prevent immobilization of analyte protein during the subsequent interaction step. The chemical structure of the binding layers forms easily-activated carboxylic groups, rendering especially high binding capacity and ligand activity. It results in more active ligand on the surface, thus higher analyte signals and higher assay sensitivity.
Different amounts of ligand may be amine-coupled to the GLC, GLM, and GLH sensor chips by controlling the amount of ligand in solution during the immobilization, by tuning the activation level, or by adjusting the length of time of the immobilization step. The amount of ligand bound is monitored by following the binding response displayed on the sensorgram in real time. However, specific applications may require a surface with a very high or low ligand surface capacity. In such cases GLH and GLC chips that have a very high or low density alginate coating may be used.
For immobilizing targets through a site-specific tag, ProteOn sensor chips such as the NLC, HTG and HTE chips can be used. The NLC chip is coated with NeutrAvidin for immobilization of biotinylated ligands and the HTG/HTE chips are coated with a unique and innovative tris-NTA surface for immobilization of histidine-tagged proteins.
Sensor chips come packaged in a sealed pouch with an inert gas, have a shelf life of two years if stored properly at 4°C, and are guaranteed for six months from the date of receipt. Sensor chips are continually monitored for quality and have excellent spot-to-spot reproducibility within the 6 x 6 array.
Read more to learn about the different sensor chip types (Figure 1) and which is best for your specific application.

| Chip Selector | |
| GLC | Compact capacity amine coupling for protein-protein interactions |
| GLM | Medium capacity amine coupling for protein-protein and protein–small molecule interactions |
| GLH | High capacity amine coupling for protein–small molecule interactions |
| NLC | NeutrAvidin for biotinylated molecule capture |
| HTG | Compact capacity tris-NTA for histidine-tagged large molecule capture |
| HTE | High capacity tris-NTA for histidine-tagged small molecule capture |
| LCP | Used with the LCP Capturing Reagent Kit for liposome capture |
Fig. 1. An overview of the seven different types of ProteOn sensor chips (GLC, GLM, GLH, NLC, HTG, HTE, and LCP) with the specific application for each chip listed.
There are seven types of ProteOn sensor chips that can be used for a variety of different immobilization strategies and the creation of different capacity surfaces. The GLC, GLM, and GLH sensor chips are designed for general amine coupling of ligands, whereas the NLC and HTG/HTE sensor chips are designed for site-specific attachment of biotinylated ligands or histidine-tagged proteins, respectively. Figure 1 shows an overview of the different sensor chip types and the specific applications in which they are used.
Amine Coupling ProteOn Sensor Chips: GLC, GLM, and GLH
Three ProteOn sensor chips are available for amine coupling. The GLC, GLM, and GLH chips provide for compact, medium, and high ligand surface capacities, respectively. All three of these chips are functionalized with easily-activated carboxylic acid groups that can be reacted with the activation reagents EDC and sulfo-NHS to react specifically with free surface amines of proteins. Bio-Rad offers the amine coupling kit for use with the GLC, GLM, and GLH sensor chips.
GLC Sensor Chip — Compact Binding Capacity
The GLC sensor chip is designed with a 2-D alginate layer for amine coupling of protein ligands at a compact
(~ 6 kRU) surface capacity. The 2-D structure of the GLC chip helps mitigate mass transfer effects that are often observed with 3-D surfaces. This versatile sensor chip is ideal for probing protein-protein interactions (Figure 2).
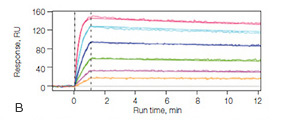
Fig. 2. A, the thin alginate coating on the GLC sensor chip responsible for creating a compact-capacity surface. B, sensorgrams of the interaction between the cytokine IL-2 and the anti–IL-2 antibody using the GLC sensor chip. The IL-2 antibody was immobilized to approximately 2,000 RU, and IL-2 was injected in a twofold dilution series ranging from 80 nM to 2.5 nM. The nearly planar surface of the GLC sensor chip allows for high-quality kinetic analysis.
GLM Sensor Chip — Medium Binding Capacity
The GLM sensor chip is coated with a thicker alginate polymer that displays a higher amount of carboxylic acid groups and is thus ideal for creating medium capacity (~ 12 kRU) ligand surfaces via amine coupling. It can be used for both protein-protein interactions and protein-small molecule interactions (Figure 3).

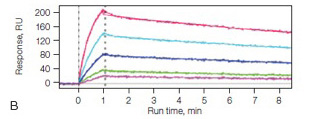
Fig. 3. A, the extended alginate coating on the GLM sensor chip responsible for creating a medium-capacity surface. B, sensorgrams of a TEM1 β-lactamase mutant interacting with the β-lactamase inhibitor protein (BLIP) using the GLM sensor chip. TEM1 was immobilized to approximately 1,500 RU, and BLIP was injected in a twofold dilution series ranging from 600 nM to 38 nM.
GLH Sensor Chip — High Binding Capacity
The GLH sensor chip is designed with a high density alginate polymer that contains an increased number of carboxylic acid groups to amine coupling ligands at a very high (>20 kRU) surface capacity. This dense alginate layer on the GLH chip is far superior at binding high capacity ligand surfaces when compared to results of ligand immobilizations done using the GLC and GLM chips (Figure 4). This sensor chip is ideal for probing protein-small molecule (<1000 daltons) interactions, as the high capacity surface gives an increased binding response. A comparison between the GLH sensor chip and a competitor's high capacity sensor chip shows the full advantage of the ProteOn chip's easily-activated carboxylic groups, rendering significantly higher binding capacity and activity, and thus much higher analyte response (Figure 5).
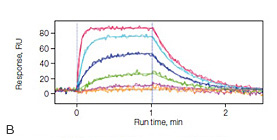
Fig. 4. A, the dense alginate coating on the GLH sensor chip responsible for creating a high-capacity surface. B, sensorgrams of the interaction between the carbonic anhydrase II (30 kD) and the inhibitor 4-carboxybenzenesulfonamide (CBS) (201 Dalton) using the GLH sensor chip. Carbonic anhydrase II was immobilized to approximately 24,000 RU, and CBS was injected in a threefold dilution series ranging from 20 µM to 0.082 µM.
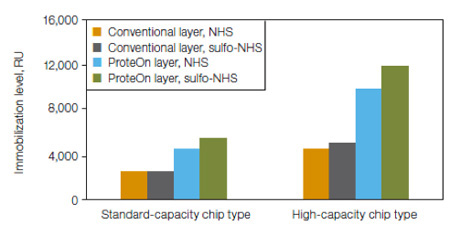
Fig. 5. Comparative coupling efficiency. Representative data for immobilization of rabbit IgG. Ligand coupling efficiency of ProteOn chip's easily activated layers is higher than conventional layers, and activation of ProteOn chip layers is higher using sulfo-NHS instead of NHS.
| Protein | pI | Non-Bio-Rad Chip, NHS Activation, RU | GLM Chip, NHS Activation, RU | GLM Chip, Sulfo-NHS Activation, RU | GLH Chip Sulfo-NHS Activation, RU |
| Pepsin | 3 | 70 | 750 | 2,050 | 2,470 |
| Ovalbumin | 4.5 | 2,800 | 3,400 | 6,700 | 6,800 |
| Protein A | 5.1 | 4,300 | 3,500 | 6,000 | 18,800 |
| β2-microglobulin | 5.3 | 2,600 | 3,250 | 3,650 | 12,400 |
| Carbonic anhydrase II | 5.9 | 6,600–2,300 | 6,000 | 9,000 | 21,200 |
| Myoglobin | 6.9–7.4 | 3,900 | 2,800 | 7,000 | 12,200 |
| Polyclonal IgG | 6–8 | 10,000 | 9,700 | 12,200 | 22,200 |
Fig. 6. Representative immobilization efficiencies on ProteOn sensor chip surfaces designed for high protein binding capacity.
ProteOn Sensor Chips for Site-Specific Attachment: NLC and HTG/HTE
NLC Sensor Chip — Immobilization of Biotinylated Ligands
The NLC sensor chip is functionalized with NeutrAvidin bound to the alginate polymer and can capture biotinylated proteins, peptides, and nucleic acids. It can capture ~ 2,000 RU of IgG or ~ 500 RU of DNA. The NLC sensor chip is ideal for immobilizing ligands without amine coupling but requires that the ligand be modified with biotin prior to immobilization (Figure 6).
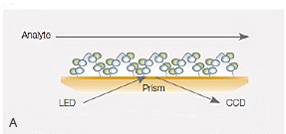
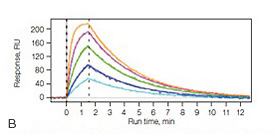
Fig. 7. A, the NeutrAvidin-modified alginate coating on the NLC sensor chip. B, sensorgrams of the interaction between an antibody Fab fragment and biotinylated MHC I/Tyr antigen using the NLC sensor chip. MHC I was captured to approximately 800 RU, and the Fab was injected in a twofold dilution series ranging from 500 nM to 31 nM.
HTE and HTG Sensor Chips — Immobilization of Histidine-Tagged Proteins
The HTG and HTE sensor chips feature a novel tris-NTA complex for improved capturing of histidine-tagged proteins. This tris-NTA complex has a significantly higher binding stability as compared to the traditional mono-NTA, so that minimal ligand leaches off the surface and the sensorgram baseline remains stable. Mono-NTA is the traditional method used to capture histidine-tagged proteins but the binding is less tight, causes ligand to leach off the surface and results in unstable baselines and distorted kinetic results, all of which can lead to an inaccurate fit to a binding model. The tris-NTA complex contains three NTA moieties for improved binding stability and increased binding selectivity to histidine-tagged proteins. The tris-NTA complex is attached to the alginate polymer matrix on the sensor chip and is activated by injecting nickel (II) ions. In order to achieve optimal performance in various applications, the surface density of tris-NTA complex is distinguished in the two different sensor chips, HTG for compact density and HTE for high density (Figure 7). The HTG and HTE sensor chips allow easy surface regeneration, chip reuse, and capture of histidine-tagged proteins directly from crude cell lysates. The HTG sensor chip is an ideal choice for protein-protein and protein-peptide interaction analysis, and the HTE sensor chip for protein–small molecule interaction analysis (Figure 8). Bio-Rad offers the HTG and HTE reagent kit for use with the HTG and HTE sensor chips.
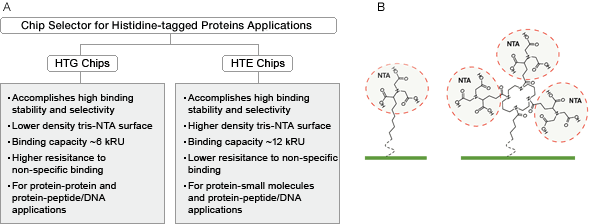
Fig. 8. A, the main features and applications of ProteOn HTG and HTE sensor chips. B, structure of surface bound mono-NTA molecule (left) and tris-NTA molecule (right). Each individual NTA group is circled.
Fig. 9. A, alginate coating modified with compact-density tris-NTA on the HTG sensor chip. B, alginate coating modified with high-density tris-NTA on the HTE sensor chip.
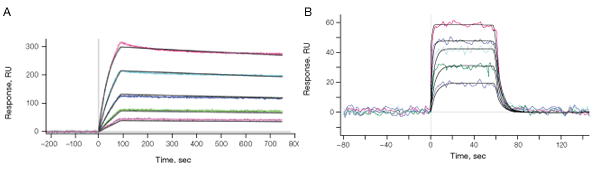
Fig. 10. A, sensorgrams of the interaction between the histidine-tagged protein A and human IgG, showing the ability of the HTG sensor chip to resolve high-affinity kinetics requiring long dissociation times. Protein A was captured to approximately 60 RU, and human IgG was injected in a twofold dilution series ranging from 100 nM to 6.3 nM. B, sensorgrams of the interaction between histidine-tagged ERK2 (an MAP kinase) and the inhibitor Purvalanol B (432.9 Dalton), showing that small molecules can be screened using the HTE sensor chip. ERK2 was captured to approximately 12,800 RU, and Purvalanol B was injected in a threefold dilution series ranging from 50 µM to 0.62 µM.
ProteOn Sensor Chips for Capturing Lipid Assemblies: Modified GLC and LCP
Biological research focused on biomolecular interactions involving lipid assemblies, such as liposomes and lipoparticles, allows the study of native membrane proteins as well as the role of the lipid bilayer of these assemblies in the activity of the membrane proteins. Analyzing lipid assemblies also provides insights into lipid-protein or lipid–small molecule interactions, answering critical questions in the fields of drug delivery, virology, and signal transduction.
Modified GLC Sensor Chip – Lipophilic Surface Chemistry
The GLC sensor chip can be modified for capturing lipid assemblies. The surface lipophilicity of the chip is adjusted through the amine coupling of an alkyl chain for capturing lipid substances. This capturing approach provides the flexibility to control the lipophilicity of the chip surface for customized lipid-based applications.
The modified GLC sensor chip provides a traditional lipophilic surface chemistry for capturing lipid assemblies, which allows for typical lipid-based applications. Bio-Rad offers the GLC lipid kit as an all-in-one kit composed of the GLC sensor chip and the lipid modification kit. The advantages of this application kit are summarized below:
- Lipophilic surface chemistry
- Flexibility in adjusting surface properties
- Good regeneration capability
- Low cost
- High throughput
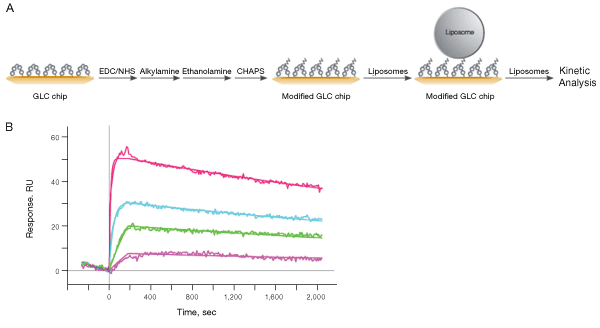
Fig. 11. A, workflow for liposome capture using the ProteOn GLC lipid kit, based on the traditional lipophilic surface chemistry. The lipophilicity of the GLC chip surface is adjusted through surface modification in order to capture lipid assemblies such as liposomes. B, detailed kinetic analysis of the interaction between CXCR4 lipoparticles captured on the modified GLC sensor chip and anti-CXCR4 antibody. CXCR4 lipoparticles were captured to approximately 3800 RU, and the anti-CXCR4 antibody was injected in a twofold dilution series ranging from 33 nM to 0.41 nM.
LCP Sensor Chip – Hydrophilic Surface Chemistry
The LCP sensor chip provides a surface functionalized with NeutrAvidin in a planar configuration that is formed on a self-assembled monolayer. It is designed to be used with the ProteOn LCP capturing reagent kit for lipid-protein, lipid–small molecule and membrane protein–protein interaction analysis. The reagent kit activates the chip surface by a biotinylated DNA tag so that the chip is able to capture DNA-labeled lipid assemblies through DNA hybridization. The reagent kit attaches DNA tags to the lipid assemblies in order to anchor them to the chip surface. It is possible to capture two or more layers of lipid assemblies for additional sensitivity. This method of capturing allows for lipid-based interaction analysis, including the analysis of membrane proteins embedded in a lipid bilayer.
The LCP sensor chip provides a novel hydrophilic surface chemistry for capturing lipid assemblies, which allows for novel lipid-based applications. Bio-Rad offers the liposome capturing kit as an all-in-one kit composed of the LCP sensor chip and the LCP capturing reagent kit. The advantages of this application kit are summarized below:
- Hydrophilic surface chemistry
- Low nonspecific binding
- Multiple layer capturing capability
- High regeneration capability
- High throughput
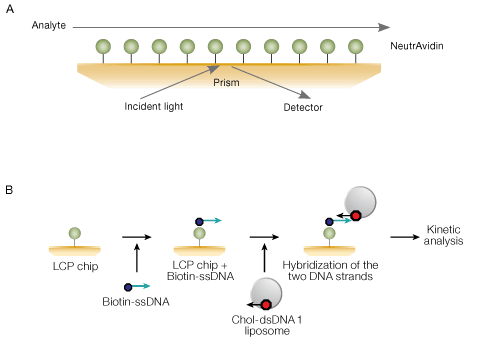
Fig. 12. A, planar NeutrAvidin-modified self-assembled monolayer on the LCP sensor chip. B, workflow for liposome capturing using the LCP chip and the LCP capturing reagent kit. Chol-dsDNA 1 and single-stranded biotinylated DNA molecules (biotin-ssDNA) contain complementary DNA sequences. The LCP sensor chip surface is saturated with biotin-ssDNA, and then liposomes incubated with chol-dsDNA 1 are captured to the surface through DNA hybridization. For reagents and techniques used in this workflow, refer to bulletin 6161.
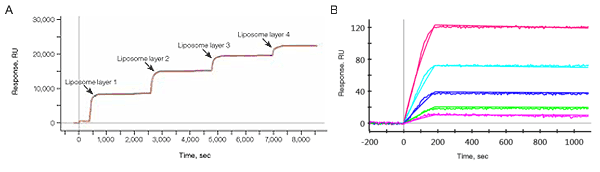
Fig. 12. A, the planar NeutrAvidin surface and the LCP capturing reagent kit allow stable capturing of several plain POPC liposome layers. B, sensorgrams of the interaction between FITC-labeled DSPC liposomes captured on the LCP sensor chip and an anti-FITC antibody. FITC-labeled DSPC liposomes were captured to approximately 330 RU, and the anti-FITC antibody was injected in a twofold dilution series ranging from 10 nM to 0.63 nM.
Which sensor chip you choose depends on a number of experimental parameters. To help you decide you may consider the following questions:
- What is the application?
- What kind of information do I want to get from my ProteOn experiment?
- What type of samples am I working with?
- Proteins?
- Peptides?
- Biotinylated or histidine-tagged proteins?
- Small molecules?
- Whole cells?
- DNA?
- Do I know anything about the affinity (tightness) of binding?
- What are the molecular weights of my sample?
For routine examination of protein-protein/protein-peptide interactions such as antibody and antigen, the GLM sensor chip is a good first choice. The moderate surface capacity allows for immobilization of ligand at surface capacities of up to 13 kRU, which is generally much higher than the surface capacity needed to obtain high quality kinetic data with protein-protein interactions. The GLC or GLH chips can be used as needed if troubleshooting your interaction with the GLM chip suggests that you need a very compact or high capacity surface. Knowing the molecular weight of your analyte is also crucial for choosing the right sensor chip, as low molecular weight analytes such as small molecules need a higher surface capacity ligand surface and will benefit from the enhanced sensitivity afforded by the GLH chip. An additional consideration to take into account when choosing a sensor chip is whether or not your interaction is mass transport limited — in other words, whether or not your interaction has a fast association rate. High capacity surfaces exacerbate a mass transport limited interaction because high amounts of ligand on the sensor chip deplete the surrounding solution of analyte very quickly, whereas compact capacity ligand surfaces mitigate the influence of mass transport by decreasing the rate at which analyte is depleted from the surrounding solution. Thus, the GLC chip with a compact capacity surface may be more suitable than the high capacity GLH chip in dealing with a mass transport limited interaction.
If your ligand contains a histidine-tag or can be biotinylated easily, the HTG/HTE and NLC sensor chips can be used to immobilize ligand to the sensor chip without needing to use amine coupling reagents. An additional benefit of using the NLC and HTG/HTE sensor chips is that the ligand is immobilized to the sensor chip surface at a specific location, as opposed to several random locations with amine coupling.
Related Content
TEST
General ProteOn Literature
| 5390 | ProteOn — PIA XPR36 Brochure | Click to download |
| 5538 | ProteOn XPR36 — Analyzing Protein Interaction Array System Featured Article Reprint | Click to download |
| 5413 | ProteOn XPR36 Hardware — 36 Interactions on a Single Chip: Label-Free, in Real-Time PIS | Click to download |
| 5627 | ProteOn Manager Software PIS | Click to download |
| 5404 | ProteOn Sensor Chips — Application-Specific Surface Chemistries = Optimized Ligand Activty PIS | Click to download |
| 5410 | ProteOn Protocol Development Kits PIS | Click to download |
| 5409 | Protein Interaction Analysis — ProteOn XPR36 System Ordering Information Sheet | Click to download |
| 6414 | ProteOn XPR36 Experimental Design and Application Guide, Rev B | Click to download |
Large Molecule Protein-Interaction Analysis
| 3172 | ProteOn PIA — Rapid and Efficient Determination of Kinetic Rate Constants using the ProteOn XPR36 Protein Interaction Array System Tech Note | Click to download |
| 5412 | ProteOn App Guide — Antibody Characterization and Development Using the ProteOn XPR36 PIA System App Guide PIS | Click to download |
| 5540 | ProteOn — PIA Screening, Ranking, and Epitope Mapping of Anti-Human IL-9 Supernatants TN | Click to download |
| 5368 | ProteOn PIA — Analysis of Multiple Protein-Protein Interactions Using the ProteOn XPR Protein Interaction Array System Tech Note | Click to download |
| 5449 | Protein Interaction Analysis Applications of the ProteOn NLC Sensor Chip: Antibody-Antigen, DNA-Protein-Protein Interactions Tech Note | Click to download |
| 5358 | ProteOn PIA — Mechanisms of Protein Binding: Double-Mutant Cycle Analysis Using the ProteOn XPR36 System Tech Note | Click to download |
| 5820 | ProteOn — Rapid Screening and Selection of Optimal Antibody Capturing Agents Using the ProteOn XPR36 Protein Interaction Array System | Click to download |
| 5360 | ProteOn PIA — Rapid and Detailed Analysis of Muliple Antigen-Antibody Pairs Using the ProteOn XPR36 Protein Interaction Array System Tech Note | Click to download |
| 5367 | ProteOn PIA — Rapid Optimization of Immobilization and Binding Conditions for Kinetic Analysis of Protein-Protein Interactions using the ProteOn XPR36 PI Array System Tech Note | Click to download |
Small Molecule Analysis
| 5965 | Rapid High-Throughput Screening of Protein Kinase Inhibitors Using the ProteOn PIA System | Click to download |
| 5797 | Protein Interaction — Rapid Assay Development and Optimization for Small Molecule Drug Discovery Tech Note | Click to download |
| 5679 | ProteOn — Applications of the ProteOn GLH Sensor Chip: Interaction Between Proteins and Small Molecules | Click to download |
| 5960 | High-Throughput Profiling of Kinase Inhibitors Selectivity Using the ProteOn XPR36 Protein Interaction Array System | Click to download |
| 5846 | Determining the Binding Kinetics of HIV-1 Nucleocapsid Protein to Six Densities of Oligonucleotide Using the ProteOn XPR36 Protein Interaction Array System TN | Click to download |
| 5822 | How to Perform Excluded Volume Correction on the ProteOn XPR36 Protein Interaction System PG | Click to download |
