The first dimension in a 2-D gel electrophoresis experiment involves the separation of proteins according to their isoelectric point (pI) by isoelectric focusing (IEF). IEF works by applying an electric field to protein within a pH gradient. The proteins separate as they migrate through the pH gradient in response to the applied voltage. When a protein reaches a pH value that matches its pI, its net electrical charge becomes neutral, and stops migrating. In this way, each protein in a sample becomes "focused" according to its pI. IEF can be performed using two techniques: immobilized pH gradients (IPG) with ampholytes covalently bound to a gel, or carrier ampholytes that migrate through a gel to generate the pH gradient. This section provides technical details to perform successful IEF using IPG strips.
Related Topics: Second-Dimension Separation, Protein Sample Preparation for 2-D Electrophoresis, Staining and Visualization of Proteins After 2-D Electrophoresis, Imaging and Analysis of 2-D Electrophoresis Gels , Protein Spot Excision and Protein Identification, and Troubleshooting Electrophoresis Gels with 2-D Doctor™.
Page Contents
Bio-Rad offers a number of products for IEF — the PROTEAN® i12™ IEF system, ReadyStrip™ IPG strips and other reagents, and accessories give the user many options for maximum flexibility.

When a protein is placed in a medium with a pH gradient and subjected to an electric field, it will initially move toward the electrode with the opposite charge. During migration through the pH gradient, the protein will either pick up or lose protons. As it migrates, its net charge and mobility will decrease and the protein will slow down. Eventually, the protein will arrive at the point where the pH gradient is equal to its pI. There, being uncharged, it will stop migrating (see figure below). If this protein should happen to diffuse to a region of lower pH, it will become protonated and be forced back toward the cathode by the electric field. If, on the other hand, it diffuses into a region of pH greater than its pI, the protein will become negatively charged and will be driven toward the anode. In this way, proteins condense, or are focused, into sharp bands in the pH gradient at their individual characteristic pI values.
Focusing is a steady-state mechanism with regard to pH. Proteins approach their respective pI values at differing rates, but remain relatively fixed at those pH values for extended periods. By contrast, proteins in conventional electrophoresis continue to move through the medium until the electric field is removed. Moreover, in IEF, proteins migrate to their steady-state positions from anywhere in the system.
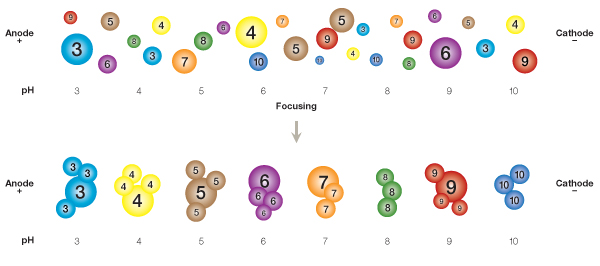
In IEF, a mixture of proteins is resolved on a pH 3–10 IPG strip according to each protein's pI and independently of its size.
A stable, linear, and reproducible pH gradient is crucial to successful IEF. IPG strips offer the advantage of gradient stability over extended focusing runs (Bjellqvist et al. 1982). IPG strips are much more difficult to cast than carrier ampholyte gels (Righetti 1983); however, IPG strips are commercially available (see ReadyStrip IPG Strips for available gradients and sizes).
pH gradients for IPG strips are created with sets of acrylamido buffers, which are derivatives of acrylamide containing both reactive double bonds and buffering groups. The general structure is CH2=CH–CO–NH–R, where R contains either a carboxyl [–COOH] or a tertiary amino group (e.g., –N(CH3)2). These acrylamide derivatives are covalently incorporated into polyacrylamide gels at the time of casting and can form almost any conceivable pH gradient (Righetti 1990).
ReadyStrip IPG Strips
A reproducible pH gradient is crucial for successful IEF. Commercially available IPG strips are easy to use and provide reproducible and stable gradients during extended IEF runs. They are cast on plastic backings, making them much easier to handle than ampholyte gels. The strip length required is dictated largely by the size of the second-dimension gels to be used, with longer strips and larger gels providing larger sample capacity and higher resolution. Bio-Rad's ReadyStrip IPG strips are produced using high-purity IPG monomers and are available in different lengths and in a variety of pH gradients
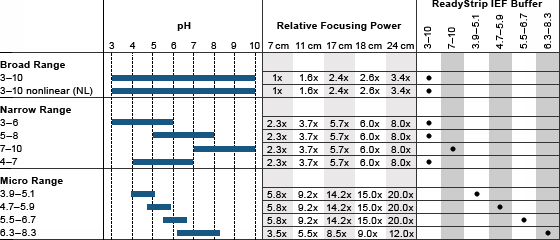
Relative focusing power of IPG strips
Relative focusing power expresses the enhanced resolution expected in the first dimension when using IPG strips of different lengths or pH ranges. The 7 cm pH 3–10 IPG strip is assigned a baseline focusing power of 1.0 to calculate the relative focusing powers of the other strips, as shown in the table above. The calculations are performed as follows:
- When two strips have the same pH range but differ in length — the ratio of the strip lengths gives the relative focusing power of the longer strip. Thus, compared to a 7 cm pH 3–10 strip, the 11 cm pH 3–10 strip has a relative focusing power of 11 cm/7 cm = 1.6
- When two strips have the same length but differ in pH range — the ratio of the pH ranges (in number of pH units) gives the relative focusing power of the strip with the smaller range. Thus, compared to a 7 cm pH 3–10 strip, the 7 cm pH 5–8 strip has a relative focusing power of (7 pH units)/(3 pH units) = 2.3
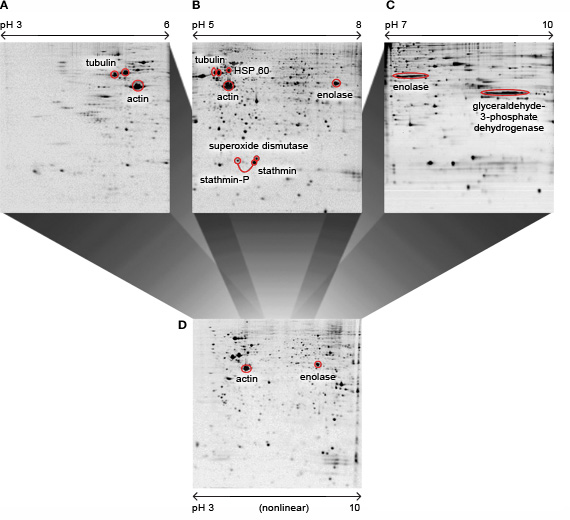
Use of broad range strips (pH 3–10) allows the display of most proteins in a single gel. With narrow range and micro range overlapping gradient strips, resolution is increased by expanding a small pH range across the entire width of a gel. Since many proteins are focused in the middle of the pH range 3–10, some researchers use nonlinear (NL) gradients to better resolve proteins in the middle of the pH range and to compress the width of the extreme pH ranges at the ends of the gradients. However, overlapping narrow range and micro range linear IPG strips can outperform a nonlinear gradient and display more spots per sample (see figure below). This result is due to the extra resolving power from use of a narrower pI range per gel. Use of overlapping gradients also allows the ability to create "cyber" or composite gels by matching spots from the overlapping regions using imaging software. Proteins that are outside of the pH range of the strip are excluded, therefore, more total protein mass can be loaded per strip, allowing more proteins to be detected. The figure below demonstrates the resolution achieved using 3 overlapping gradients with 17cm ReadyStrip IPG strips and large format gels.
Narrow overlapping IPG strips outperform nonlinear IPG strips. Proteins were separated by 2-D PAGE using 17 cm IPG strips, then stained with Coomassie Blue. A, 2 mg protein on pH 3–6 ReadyStrip IPG; B, 2 mg protein on pH 5–8 ReadyStrip IPG; C, 1 mg protein on pH 7–10 ReadyStrip IPG; D, 0.8 mg protein on a nonlinear pH 3–10 IPG. With the narrow range strips (A–C), more sample was loaded, and consequently, more spots were resolved and detected after staining compared to the wide-range IPG strip (D). Kindly provided by Sjouke Hoving, Hans Voshol, and Jan van Oostrom of Novartis Pharma AG, Functional Genomics Area, Basel, Switzerland.
The 17, 18, and 24 cm IPG strips and large format gels have a large area to resolve proteins spots; however, they take a long time to run. Using a mini system instead of, or as a complement to, a large gel format can provide significant time savings. A mini system, such as Bio-Rad's Mini-PROTEAN® system is perfect for rapid optimization of sample preparation methods. Switching to a large format then allows thorough assessment of a complex sample and identification of proteins of interest. In many cases, a mini system consisting of narrow range IPG strips can then be used to focus in on the proteins of interest.
Throughput of the 2-D process is a consideration when choosing gel size. The table below compares gel size, run times, and equipment. The ability to cast or run 12 gels at a time in any 3 size formats is very useful in gathering proteomic results. In some cases, mini systems (7cm ReadyStrip IPG strips with Mini-PROTEAN® precast gels, or 11cm ReadyStrip IPG strips with Criterion™ precast gels) can completely replace large 2-D systems, providing speed, convenience, and ease in handling. The availability of narrow and micro overlapping pH-range ReadyStrip IPG strips can increase the effective width of pI resolution more than 5-fold after accounting for overlapping regions. When 3 narrow range overlapping ReadyStrip IPG strips are used with the Criterion system, the resolution in the first dimension is increased from 11 to 26 cm. When micro-range strips are used, the resolution in the first dimension is expanded from 11 to 44 cm.
2-D Dimension Instrument Selection Chart
Use the chart below to find the most suitable second dimension system that best meets your needs based on IPG strip length, gel dimensions, and number of gels that can be run at once.
| First Dimension IEF | Second-Dimension Electrophoresis | ||||
| Focusing Cell | IPG Length | Vertical Electrophoresis Cell | # Gels/Cell | Gel Dimension | Run Time |
 PROTEAN® i12™ IEF system |
18 or 24 cm |  PROTEAN® Plus Dodeca™ cell |
12 | 25 x 20.5 cm 20 x 20.5 cm 25 x 20.5 cm |
6 hr |
| 17 cm |  PROTEAN® II XL cell |
2 | 18.3 x 19.3 cm | 4–6 hr | |
| 17 cm |  PROTEAN® II XL Multi-cell |
6 | 18.3 x 20 cm 18.3 x 19.3 cm |
4–6 hr | |
| 11 cm |  Criterion™ cell |
2 | 13.3 x 8.7 cm | 30–60 min | |
| 11 cm |  Criterion™ Dodeca cell |
12 | 8.6 x 6.8 cm | 60 min | |
| 7 cm |  Mini-PROTEAN® Tetra cell |
4 | 8.6 x 6.8 cm | 15–45 min | |
| 7 cm |  Mini-PROTEAN® 3 Dodeca cell |
12 | 8.6 x 6.8 cm | 35 min | |
IPG strips are dehydrated and must be rehydrated to their original gel thickness (0.5 mm) before use. This allows flexibility in applying sample to the strips. There are 3 methods for sample loading: passive in-gel rehydration with sample, active in-gel rehydration with sample, or cup loading of sample after IPG rehydration. The easiest and most efficient way of applying the sample is while rehydrating the strip. In some specific instances, it is best to rehydrate the strips and then apply sample through sample cups while current is applied.
Sample Application During Rehydration
For both active and passive rehydration methods, the sample is introduced to the IPG strip at the time of rehydration. As the strips hydrate, proteins in the sample are absorbed and distributed over the enter length of the strip (Sanchez et al. 1997).
- Active Rehydration — a very low voltage is applied during rehydration of the strips. Proteins enter the gel matrix under current as well as by absorption. Active rehydration is thought to help large proteins enter the strip by applying electrical "pull". Because the voltage is applied before all the solution and proteins are absorbed into the gel, the pH of a protein's environment will be the pH of the rehydration buffer, and the protein will move according to its mass-to-charge ratio in that environment. Thus, small proteins with a higher mobility have a higher risk of being lost from the strip.
- Passive Rehydration — proteins enter the gel by absorption only. This method allows efficient use of equipment since strips can be rehydrated in sample rehydration trays while other samples are being focused in the IEF cell.
Whether the strips are hydrated actively or passively, it is very important that they be incubated with sample for at least 12 hr prior to focusing. This allows the high molecular weight proteins time to enter the gel after the gel has become fully hydrated and the pores have attained full size. These sample application methods work because IEF is a stead-state technique, so proteins migrate to their pI independent of their initial positions.
The advantages of this approach are:
- Sample application is simple (Gorg et al. 1999)
- Sample application during rehydration reduces the risk of sample precipitation, which often occurs with cup loading (Rabilloud 1999)
- Shorter focusing times can be used because the sample proteins are in the IPG strip prior to IEF
- Very large amounts of protein can be loaded using this method
Sample Application By Cup Loading
When loading the protein sample from a cup, the IPG strips must be rehydrated prior to sample application. In general, focusing is most effective towards the end of the strip opposite the site of the sample cup placement. Use anodic sample cup placement when using basic pH ranges or when resolution of basic proteins is desired.
Cup loading can be beneficial in the following cases:
- When samples contain high levels of DNA, RNA, or other large molecules, such as cellulose
- For analytical serum samples that have not been treated to remove albumin
- When running basic IPG strips; e.g., pH 7–10
- For sample that contain high concentrations of glycoproteins
During an IEF run, the electrical conductivity of the gel changes with time, especially during the early phase. When an electric field is applied to an IPG at the beginning of an IEF run, the current will be relatively high due to the large number of charge carriers present. As the proteins and ampholytes move toward their pIs, the current will gradually decrease due to the decrease in the charge on individual proteins and carrier ampholytes.
The pH gradient, strip length, and the applied electric field determine the resolution of an IEF run. According to both theory and experiments, the difference in pI between two adjacent IEF-resolved protein bands is directly proportional to the square root of the pH gradient and inversely proportional to the square root of the voltage gradient at the position of the bands (Garfin 2000).
Thus, narrow pH ranges and high applied voltages yield higher resolution in IEF. The highest resolution can be achieved using micro-range IPG strips and an electrophoretic cell, such as the PROTEAN i12 IEF system, capable of applying high voltages. IEF runs should always be carried out at the highest voltage compatible with the IPG strips and electrophoretic cell. At the completion of focusing, the current drops to nearly zero since the carriers of the current have stopped moving. The PROTEAN i12 IEF system is designed to provide precise cooling, allowing the highest possible voltages to be applied.
The PROTEAN i12 IEF system offers individual lane control allowing running different samples, pH gradients and protocols to be run simultaneously. A traditional IEF cell sets a current limit that is averaged among the lanes. If one sample has higher conductivity, it will pull more current which can result in over-focusing or even burning the IPG strip. This can also result in under-focusing of the lower conductivity sample resulting in a streaky 2D gel. It is recommended when running traditional IEF cells that the samples are very similar in conductivity and the pH ranges of the strips be identical to avoid these problems. With the PROTEAN i12 IEF system, each lane is controlled independently and the current limit can be set for each lane. A current limit for each sample can be reliably set and will not vary based on the other samples in the experiment. This will result in more consistent and reproducible results. A default current limit of 50 µA per strip is intended to minimize protein carbamylation reactions in urea sample buffers. This limit can be increased to 100 µA per strip if desired.
Given that the pH gradient is fixed in the IPG strip gel, focused proteins are more stable at their pI than in conventional IEF gels. Focused IPG strips can be stored at –20°C indefinitely without affecting the final 2-D pattern. IPG strips are bound to a plastic sheet, so gel cracking, which results from expansion and contraction during freezing and thawing, is avoided and the IPG strips retain their original dimensions after thawing. It is convenient to store IPG strips in rehydration trays, which can then be used to equilibrate the strips for the second dimension.
Videos
Documents
TEST
| Number | Description | Options |
|---|---|---|
| 6240 | First-Dimension Separation Methods | Click to download |