Nucleic acid analysis generally involves isolation and characterization of the DNA or RNA sample of interest. Sample purification and quality assessment are important steps in experimental workflows since the quality of the recovered nucleic acid can affect the performance in downstream reactions. A wide range of techniques can be used to transform a sample which cannot be directly analyzed into one that fits the requirements of the analytical technique to be used.
Page Contents
DNA and RNA are nucleic acid molecules that are used to store and transmit genetic information inside a cell. The central dogma of molecular biology states that DNA is transcribed into RNA that is then translated into protein. While this is a simplistic look at gene and protein expression, it illustrates that DNA and RNA can provide important information about genomics, gene expression and cellular regulation for a sample of interest. In order to perform analysis on DNA and RNA, it is often necessary to first extract these molecules from cells or tissues.
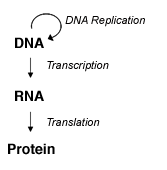
Central dogma of molecular biology.
Several well established methods exist for extracting, purifying, and concentrating nucleic acids. Selecting the most appropriate nucleic acid isolation method depends on several factors including the starting sample material, the quantity, size and stability of the target nucleic acid, and the downstream application for the nucleic acid of interest. Following the purification step, it is often necessary to determine nucleic acid concentration, purity, and integrity before proceeding with downstream applications.
Common Nucleic Acid Sample Preparation Methods
| Cesium Chloride Gradients | Filter Column Purification | Gel Extraction | Guadinidium Isothiocyanate Phenol Chloroform Extraction - Ethanol Precipitation | Ion Exchange Resin | Phenol Chloroform Extraction - Ethanol Precipitation | Size Exclusion | |
| Recommended For | Plasmid DNA | DNA and RNA > 200bp | DNA | RNA | DNA | RNA | DNA and RNA |
| Pros | Highly pure DNA | Fast and Easy | Fast and Easy | Scalable by volume: highly pure RNA | Scalable by volume | Scalable by volume: highly pure RNA | Fast and Easy |
| Cons | Requires expensive instruments and removal of ethidium bromide and cesium chloride from isolated plasmid | May not recover molecules <200 bp | Potentially low yield; agarose contamin-ation may inhibit downstream reactions | Phenol is hazardous | Residual resin may inhibit downstream reactions | Phenol is hazardous | Potentially low yield; imperfect size separation |
Gel electrophoresis can be used to effectively separate nucleic acids based on their size within agarose or polyacrylamide gels. You can successfuly separate nucleic acid fragments ranging in length from 20 base pairs to 20 kb by electrophoresis. Size ladders containing a range of different-length DNA fragments are run with the sample of interest to identify the corresponding size bands in a gel. Agarose gels are ideal for the separation of DNA restriction digests, PCR products, and genomic DNA or RNA prior to Southern or northern blotting. Polyacrylamide gels can be used for what is known as polyacrylamide gel electrophoresis, or PAGE. Applications include oligonucleotide analysis, RNase protection assays, and northern blotting. Non-denaturing polyacrylamide gels are used to separate small double-stranded DNA (dsDNA) fragments, particularly PCR products. Denaturing polyacrylamide gels are used for separation of small RNA and single-stranded DNA (ssDNA) fragments. Gels can be hand cast in the lab, or precast gels can be purchased from commercial providers.
Good laboratory technique is essential when working with nucleic acids to minimize nuclease contamination. One critical consideration when working with nucleic acid is to avoid nucleases in your solutions, consumables, and labware. Ready-to-use solutions and consumables that are nuclease-free, such as PCR-grade water, are readily available from commercial providers. Alternatively, for RNase removal, you can treat solutions with diethyl pyrocarbonate (DEPC), and then autoclave them. RNases on labware can also be inactivated by DEPC treatment, or by baking at 250°C for 3 hr.
When performing organic extractions with phenol, be aware that phenol causes burns and can be fatal if ingested. When working with phenol, use gloves and eye protection (laboratory glasses, shield, and safety goggles). Do not get on skin or clothing and avoid breathing vapor.
