Reducing the complexity of the sample prior to loading on a 2-D gel can dramatically improve the range of detection and increase the visibility of minor proteins. Techniques such as differential extraction, subcellular fractionation, chromatography, or prefocusing in a preparative IEF device such as Bio-Rad's Rotofor System have been used to reduce the complexity of samples. This section describes fractionation and depletion techniques and provides a list of Bio-Rad products that can be used in these techniques.
Related Topics: Protein Solubilization and Protein Extraction and Cleanup.
Page Contents
Below is a graphic representation to illustrate the difference between fractionation and depletion methods to enrich specific types of proteins. Fractionation reduces the complexity of a protein sample by separating proteins in different classes based on biophysical properties of the proteins (size, charge, cellular location, solubility etc.). Depletion reduces the complexity of the sample by removing abundant proteins and thereby increasing the representation of the less abundant ones.
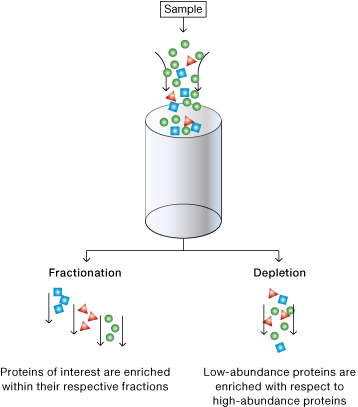
Differences between fractionation and depletion methods.
Bio-Rad's protein sample preparation products may be used with virtually all sample types. These products are based on well-understood purification and fractionation principles and may be applied individually or in combination for effective sample preparation.
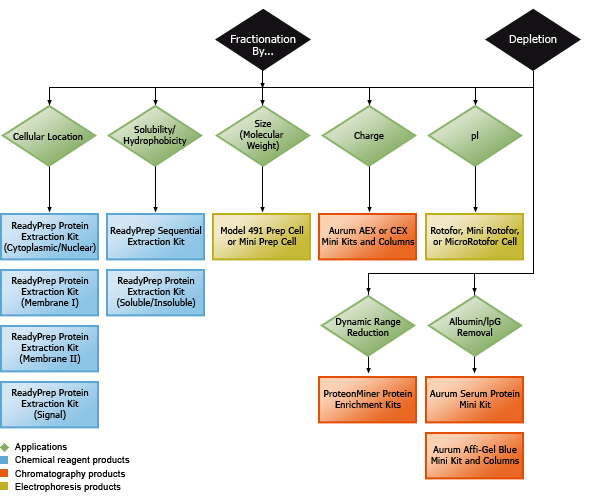
Product selection guide.
For effective 2-D analysis, it may be necessary to reduce the complexity of the protein sample, especially when analysis of low-abundance proteins is the goal. This can be achieved by separating proteins based on various criteria such as cellular location or physicochemical properties of the proteins.
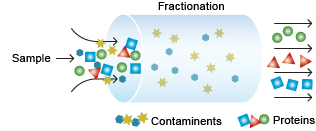
Separation of protein samples.
Cellular Location
Kits are available for the extraction of cytoplasmic and nuclear proteins (ReadyPrep™ protein extraction kit (cytoplasmic/nuclear), membrane proteins of various complexities (ReadyPrep™ protein extraction kit (membrane I) and ReadyPrep™ protein extraction kit (membrane II)), and proteins involved in intracellular membrane trafficking and signaling pathways (ReadyPrep™ protein extraction kit (signal) and ReadyPrep™ protein extraction kit (soluble/insoluble).
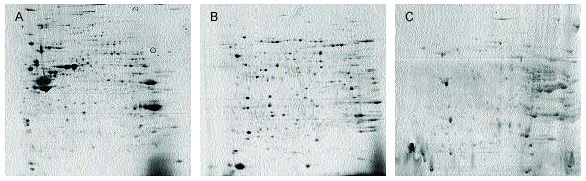
Differences in 2-D patterns obtained using ReadyPrep signal (A), membrane I (B), and membrane II (C) kits. Mouse liver samples were extracted using the recommended protocols for each kit. Purified proteins (~450 µg) were focused on 17 cm pH 3–10 nonlinear ReadyStrip IPG strips and run on 8–16% PROTEAN® II precast gels for the second dimension. Overall spot patterns differ for A, B, and C even though all three kits isolate membrane proteins, indicating each kit isolates different types of membrane proteins.
Solubility/Hydrophobicity
Available kits use differential solubility for extraction. The ReadyPrep™ sequential extraction kit uses a series of three extraction buffers, each differing in their detergent and chaotrope concentrations. The insoluble residues from each extraction step are further extracted with reagents of increasing solubilizing power.
The ReadyPrep protein extraction kit uses one step with a strong detergent to generate a soluble fraction containing hydrophilic proteins, and an insoluble fraction containing hydrophobic proteins. Proteins in the insoluble fraction are eventually resolubilized in ReadyPrep™ 2-D rehydration/solubilization buffer 1, a strongly chaotropic solution containing the zwitterionic detergent ASB-14. This simple fractionation strategy results in less complex samples, leads to improved identification of low-abundance proteins, and a better overall view of the proteome.

Distribution of proteins based on differential solubility using the ReadyPrep sequential extraction kit. The generation of three fractions provides increased resolution of proteins on 2-D gels.
Protein Size
Fractionation by size (molecular weight) is an effective enrichment strategy for studies of protein families and post-translational modifications because the sizes of these proteins tend to be similar. Products such as the Model 491 prep cell and mini prep cell use continuous elution electrophoresis to purify proteins and other macromolecules based on their relative mobility in polyacrylamide or agarose gels.

Model 491 prep cell and mini prep cell.
Protein Charge
Fractionation by charge allows separation of acidic and basic proteins. Columns such as Aurum™ ion exchange (AEX and CEX) mini columns allow selective purification of either acidic or basic proteins.

Fractionation of rat brain tissue using Aurum ion exchange mini columns. Rat brain total protein extracts (3 ml) were loaded onto an Aurum AEX or CEX mini column and eluted. The unfractionated and fractionated samples were then treated with the ReadyPrep reduction-alkylation and 2-D cleanup kits and separated by 2-D gel electrophoresis.
Protein Isoelectric Point (pI)
Protein fractionation by pI improves downstream sample loading and separation on narrow- and micro-range IPG strips by eliminating proteins outside the pH region of interest. Products such as the Rotofor, mini Rotofor, and MicroRotofor cells fractionate proteins by preparative liquid-phase IEF.
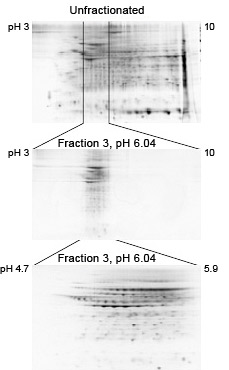
Clean fractionation by pI. Mouse liver extract was fractionated using the MicroRotofor cell. 2-D separations of the unfractionated sample (120 µg protein) and fraction 3 (20 µg protein) are shown. Prior to 2-D separation, samples were treated with the ReadyPrep 2-D cleanup kit. Note the clean pH boundaries of fraction 3 and the enrichment of proteins in the pH region it covers.
Rotofor and Mini Rotofor Cells
The Rotofor and mini Rotofor cells fractionate proteins by preparative liquid-phase IEF. Proteins (up to 1 g) are separated and concentrated into 20 discrete fractions that can be further separated by 2-D gel electrophoresis.
MicroRotofor Cells
Based on Rotofor cell technology, the compact MicroRotofor system performs liquid-phase IEF with a 2.5 ml sample capacity and generates 10 discrete fractions.

MicroRotofor Cell.
The isolation of lower-abundance proteins from serum or plasma is often complicated by the presence of albumin and immunoglobulin G (IgG). Albumin is the most abundant protein, representing 60–70% of all proteins in serum, and IgG is the second most abundant protein with a 10–20% representation. These two proteins effectively mask the presence of many comigrating proteins and limit the amount of total serum protein that can be resolved on a 2-D gel. Immunodepletion or affinity chromatography can be used to deplete high-abundance proteins. Other kits use a bead-based library of combinatorial peptide ligands to reduce levels of high-abundance proteins.
Albumin/IgG Removal
Proteomic analysis of serum presents its own unique challenges. Albumin and IgG contribute up to 90% of the total protein in a serum sample. These proteins obscure lower-abundance proteins and limit loading capacity on 2-D gels (see figure below). To obtain meaningful results, these proteins must be removed prior to downstream separation or analysis. Kits such as Aurum™ Affi-Gel® Blue and Aurum serum protein mini columns and kits can be used for the removal of both albumin and IgG. Alternatively these proteins can be removed using specific antibodies.
Before

After

Albumin and IgG removal from serum using the Aurum serum protein mini kit. Serum proteins were electrophoresed before and after treatment with an Aurum serum protein mini column. Albumin and lgG are removed following treatment with an Aurum serum protein column, improving resolution of other protein spots. Samples (100 µg protein) were focused on 11 cm ReadyStrip pH 5–8 IPG strips, then run on Criterion™ 8–16% Tris-HCl precast gels for the second dimension.
Dynamic Range Reduction
Removal of high-abundance proteins leads to enrichment of proteins of lower abundance. ProteoMiner™ technology is based on the interaction of proteins with a large, highly diverse library of hexapeptides bound to chromatographic supports (see figure below). The ProteoMiner™ protein enrichment kits can be used for enriching medium- and low-abundance proteins and for decreasing the amount of high-abundance proteins. This method is not dependent on a predefined set of antibodies. Unlike immunodepletion products, it can be applied to protein samples from a variety of sources.
Before
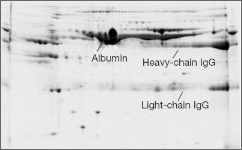
After

Enrichment of low-abundance serum proteins with the ProteoMiner protein enrichment kit. Resolution is dramatically improved on a gel with an equal amount of total protein from sample treated with ProteoMiner beads. The high-abundance proteins are greatly reduced and a greater number of protein spots are visualized. Samples (100 µg protein) were focused on 11 cm ReadyStrip pH 5–8 IPG strips, then run on Criterion 8–16% Tris-HCI precast gels for the second dimension.
Related Content
TEST
| Number | Description | Options |
|---|---|---|
| 6200 | Sample Quantitation (RC DC Protein Assay) Protocol and Compatibility Table | Click to download |
| 6221 | Sample Quantitation (RC DC Protein Assay) | Click to download |
| 6220 | Solubilization | Click to download |
