A well-designed and optimized PCR assay provides the highest specificity and yield. Special considerations should be made when performing fast PCR assays to increase throughput and when amplifying long templates. This section covers the design and optimization of an end-point PCR assay. Additional considerations are needed for the design of real-time PCR assays.
Related Topics: PCR Instrumentation, PCR Reagents, PCR Analysis, and PCR Troubleshooting.
Page Contents
A successful PCR assay requires efficient and specific amplification of the product. Both the primers and the target sequence can affect the efficiency, specificity, and accuracy of PCR assays. Therefore, care must be taken when choosing a target sequence and designing primers. The use of PCR primers specifically designed and validated for PCR assays with your target of interest is highly recommended.
When designing primers for a PCR assay, follow these steps:
- Check the literature and databases for existing primers.
- Choose a target sequence.
- Design primers.
- Check primer specificity.
- Assess primer properties (melting temperature [Tm], secondary structure, complementarity).
- Determine PCR product properties
- Optimize the protocol.
A number of free online resources are available to help you with PCR assay design (see Free Internet Resources for Primer Design). Commercially available programs such as Beacon Designer software can perform both primer design and target sequence selection.
Choosing a Target Sequence
When amplifying any sequence in a given section of DNA for purposes such as genotyping experiments, follow these guidelines to select a product. See the guidelines under Long PCR Assays when amplifying long sequences.
- Plan to amplify a 75–200 bp product. Although short PCR products are typically amplified with higher efficiency than longer ones, a PCR product should be at least 75 bp to easily distinguish it from any primer-dimers that could potentially form
- When possible, avoid regions that have secondary structure. Use programs such as mfold to predict whether a PCR product will form any secondary structure at the annealing temperature. See Real-Time PCR: General Considerations (Bio-Rad bulletin 2593) for more details
- Avoid regions with long (>4) repeats of single bases
- Choose a region that has a GC content of 50–60%
Designing Primers
When designing primers for a PCR assay, follow these guidelines:
- Design primers that have a GC content of 50–60%
- Strive for a Tm between 50 and 65°C. One way to calculate Tm values is by using the nearest-neighbor method (use this online Tm calculator)
- Avoid secondary structure; adjust primer locations so that they are located outside secondary structure in the target sequence, if required
- Avoid repeats of Gs or Cs longer than 3 bases
- Place Gs and Cs on ends of primers
- Check the sequence of forward and reverse primers to ensure no 3' complementarity (avoid primer-dimer formation)
- Verify specificity using tools such as the (Basic Local Alignment Search Tool BLAST)
Optimizing the annealing temperature of your PCR assay is one of the most critical parameters for reaction specificity. Setting the annealing temperature too low may lead to amplification of nonspecific PCR products. On the other hand, setting the annealing temperature too high may reduce the yield of a desired PCR product. Even after calculating the Tm of a primer, you may need to determine the annealing temperature empirically. This involves repeating a reaction at many different temperatures. Similar time-consuming tests may also be required to optimize the denaturation temperature.
The optimal annealing temperature for an assay can be easily determined using PCR instruments that have a thermal gradient feature. All Bio-Rad thermal cyclers offer a gradient feature. The gradient feature allows you to test a range of temperatures simultaneously, optimizing the annealing temperature in a single experiment.
To find the optimal annealing temperature for your reaction, test a range of temperatures above and below the calculated Tm of the primers. Analyze the results using agarose gel electrophoresis. If nonspecific amplification has occurred, additional bands will appear on the gel. The optimal annealing temperature is the one that results in the highest yield with no nonspecific amplification. A sample annealing temperature optimization experiment is shown in Figure 1.
If satisfactory results are not obtained at any annealing temperature, additional optimization steps must be taken, and primer redesign may be necessary. Consult the PCR Assay Troubleshooting page for more help.
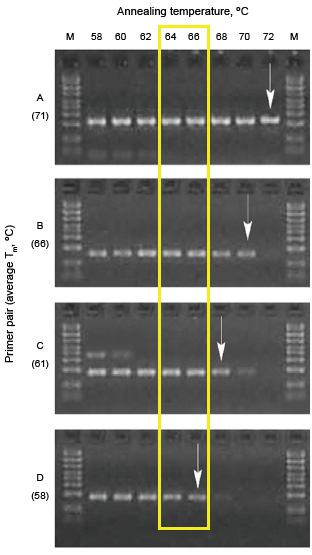
Gradient optimization of a PCR assay. All reactions were evaluated in a single run. Four different primer sets (A, B, C, and D) were designed and tested for amplification. Arrows indicate the annealing temperature that provided the highest specificity while maintaining good yield. Yellow box indicates optimal temperatures. M, markers; Tm, melting temperature.
Several advances in PCR have dramatically reduced the duration of PCR amplification reactions. Optimization of primers and protocols, the use of highly processive polymerases, and fast thermal cyclers are enabling researchers to obtain highly specific amplifications in short time periods. Optimized fast protocols can shorten PCR reactions by approximately 1 hr.
Shortening PCR Run Times
There are several ways one can shorten PCR run times. The most dramatic reduction in run time can be achieved by using a modified protocol with enzymes and supermixes that are compatible with faster reaction times. DNA polymerases have been engineered to have increased processivity and speed. Furthermore, hot-start enzymes have been modified to require less time for temperature-mediated activation. Thermal cyclers with faster ramp speeds also contribute to fast PCR, although differences in thermal cycler ramp rates have a smaller impact compared to protocol modification.
Modifying a PCR Protocol for Faster Run Times
There are several ways to modify a PCR protocol for faster run times. To modify your protocol for faster cycling of targets <250 bp:
- Select primers with higher annealing temperatures (such as 65–70°C, as calculated using Santa Lucia nearest-neighbor thermodynamic values)
- Choose a hot start enzyme that requires a minimum activation time (such as an antibody-mediated hot-start enzyme)
- Optimize a lower denaturation temperature
- Combine annealing and extension steps into a single step
Gene cloning and other downstream applications often require accurate amplification of long stretches of DNA. Successful amplification of nucleic acid regions greater than 3 kb require special considerations when designing a long PCR assay.
Amplifying long stretches of DNA provides additional challenges because of the increased probability of base misincorporation. Introduction of base-pair errors during strand elongation dramatically slows or completely halts DNA polymerization. The low fidelity of Taq polymerase renders this enzyme unsuitable for most long PCR amplifications. Employing DNA polymerases with proofreading capabilities in long PCR assays greatly increases amplification efficiencies. Proofreading enzymes have 3' → 5' exonuclease activity, allowing the enzyme to remove a misincorporated base and continue elongation. The relatively low processivity of traditional proofreading enzymes has led researchers to use Taq and proofreading polymerases in conjunction or use engineered proofreading polymerases that deliver increased polymerization rates.
The quality of DNA template also plays an important role in long PCR assays. Exposure of the template to high temperatures can lead to DNA depurination (the removal of adenine or guanine from the deoxyribose sugar on a DNA base). The length of templates used in long PCR increases the chances of depurination. The probability of DNA depurination can be decreased by shortening the heated incubation for the denaturing step to approximately 10 sec.
Long template DNA also has a greater tendency to form secondary structures that can inhibit amplification. Careful optimization of dimethyl sulfoxide (DMSO) concentrations can help alleviate these secondary structures by disrupting DNA base pairing.
For more information about long PCR assays, see Long Inverse PCR Using iProof™ Polymerase (Bio-Rad bulletin 5337).
Checking for Existing PCR Primers
- Real Time PCR Primer Sets
- PrimerBank (Massachusetts General Hospital)
- RTPrimerDB
- Quantitative PCR Primer Database (QPPD; NCI)
Choosing a Target Sequence
- Entrez Gene (NCBI)
- Ensembl Genome Browser (Sanger Institute/European Bioinformatics Institute)
- Sequence Server (Dolan DNA Learning Center)
Designing Primers
- Primer3 (Whitehead Institute for Biomedical Research, MIT)
- GeneFisher2 (Bielefeld University)
- FastPCR (PrimerDigital, Helsinki, Finland)
- PerlPrimer (Owen Marshall)
- Primer Design Assistant (Division of Biostatistics and Bioinformatics, NHRI)
- Beacon Designer (PREMIER Biosoft International)*
Designing Primers for Reverse-Transcription PCR (RT-PCR)
Checking Primer Specificity
Assessing Primer Properties
- OligoAnalyzer 3.1 (Integrated DNA Technologies)
- NetPrimer (PREMIER Biosoft International)
- Gene Walker (CyberGene AB)
- Oligo Calc: Oligonucleotide Properties Calculator (Northwestern University)
Assessing PCR Product Properties
* Indicates commercial software.
