Better Sample Preparation
Good results start with good samples.

On This Page |
Sample Preparation Basics | Cell Lysis & Fractionation | Protein Solubilization & Stabilization | Sample Quantitation | Loading Buffer | Sample Prep Tips | Protocols & Resources |
Sign Up for Western Blotting Tips and Webinar Alerts
Sample Preparation Basics
Sample preparation for protein electrophoresis involves the extraction and solubilization of a protein sample from its cellular matrix, removal of contaminants, and adjustment of total protein concentration to a suitable range. The quality of sample preparation can greatly affect electrophoresis results and the ultimate quality of the western blot data.
Sample Preparation Overview
Cell Lysis and Fractionation
For effective cell lysis that yields your protein of interest in a soluble form, there are a few factors to consider:
- First, consider the method of cell disruption as different sample types require different methods for efficient cell lysis
- Second, consider the subcellular location of your protein of interest and choose a method that enriches for that cellular compartment
- Lastly, the appropriate choice of lysis buffer can yield higher amounts of your target protein as the detergents in the buffer can affect lysis efficiency and solubilization of different proteins
Common Cell Lysis Methods

Detergent Disruption
Simple exposure to detergents can lyse cells that disrupt easily, like blood cells or tissue culture cells. Non-ionic detergents like NP-40 are mild and nondenaturing but do not solubilize hydrophobic proteins well. Zwitterionic detergents like CHAPS and ionic detergents like SDS are more effective at solubilization and denature proteins during lysis.

Mechanical Methods
Involve exposing cells to physical forces using dounces, sonicators, or grinding of tissue. Some methods like sonication generate heat which can overheat the sample so use cooling to avoid overheating. During sonication, immersion in an ice bath is sufficient. For grinding with mortar and pestle, addition of splashes of liquid nitrogen will keep the sample cool. Often both chemical disruption and some sort of mechanical method are used together to maximize protein yields.

Enzymatic Treatments
Enzymes that digest cell walls of plants or bacteria aid in cell lysis. For example, cellulase and pectinase for plant cells, lyticase for yeast cells, and lysozyme for bacterial cells. Enzyme treatment is usually followed by another disruption method such as sonication.
-
Tip
Regardless of cell disruption method, remove any insoluble material to avoid blocking the pores of the electrophoresis gel. Centrifuge extracts extensively (20,000 x g for 15 minutes at 15°C).
Techniques for Specific Sample Types
Suitability of cell disruption methods to various sample types.
| Technique | Description |
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|
| Gentle Methods | |||||||
| Osmotic lysis | Suspension of cells in hypotonic solution; cells swell and burst, releasing cellular contents |
|
|
|
|
|
|
| Freeze-thaw lysis | Freezing in liquid nitrogen and subsequent thawing of cells |
|
|
|
|
|
|
| Enzymatic lysis | Suspension of cells in iso-osmotic solutions containing enzymes that digest cell wall of plants, yeast, or bacteria. Usually followed by another disruption method, such as sonication |
|
|
|
|
|
|
| Harsher Methods | |||||||
| Sonication | Disruption of cell suspension by short bursts of ultrasonic waves. Cool on ice to avoid overheating |
|
|
|
|
|
|
| French press | Application of shear forces by forcing a cell suspension through a small orifice at high pressure |
|
|
|
|
|
|
| Grinding | Grinding of sample to fine powder with mortar and pestle. Mortar usually filled with liquid nitrogen during grinding to keep sample cold |
|
|
|
|
|
|
| Mechanical homogenization | Homogenization with a Dounce or blender. Best suited for soft, solid tissues |
|
|
|
|
|
|
| Glass-bead homogenization | Application of gentle abrasion by vortexing cells with glass beads |
|
|
|
|
|
|
Cell Lysis Volume Recommendations
| Type of cells | Amount of Material | Volume of lysis buffer |
|---|---|---|
| Tissue Culture | 107 cells or 100 mm dish |
1 ml |
| Whole Tissue | 100 mg | Add 2 ml and sonicate or dounce homogenize |
| Bacteria | Spin sample, estimate volume | Add 10 volumes and vortex |
| Yeast | Spin sample, estimate volume | Add 10 volumes, then sonicate or vortex with glass beads |
See Our Protein Extraction Products »
Cell Fractionation
Often times, the protein of interest is present at low levels or present in tissues that contain many other proteins making detection difficult. In such cases, one can employ cellular fractionation to isolate specific cell organelles to achieve more optimal detection on western blot. Subcellular fractionation can be achieved by differential centrifugation and in some instances, by using specific lysis detergents.
As an example, proteins that are known to be hydrophobic (membrane bound) can be separated from hydrophilic proteins (cytoplasmic) by incubation of cells with non-ionic detergents (Triton-X or Tween 20) which results in the formation of two distinct layers; a hydrophobic and a hydrophilic. Alternative detergents can be employed to target different subcellular components (see below).

Lysis Buffer Recommendations
| Cellular Location | Recommended Buffer |
|---|---|
| Whole cell lysate | NP-40 |
| Nuclear | RIPA, or nuclear fractionation for increased protein of interest concentration |
| Mitochondria | RIPA, or mitochondrial fractionation for increased protein of interest concentration |
| Cytoplasmic | Tris-HCl |
| Membrane-bound proteins | RIPA, (SDS is generally considered harsh and thus is often well-suited for difficult to solubilize proteins) |
|
For more details on detergents see MATER METHODS 2013;3:163 |
|
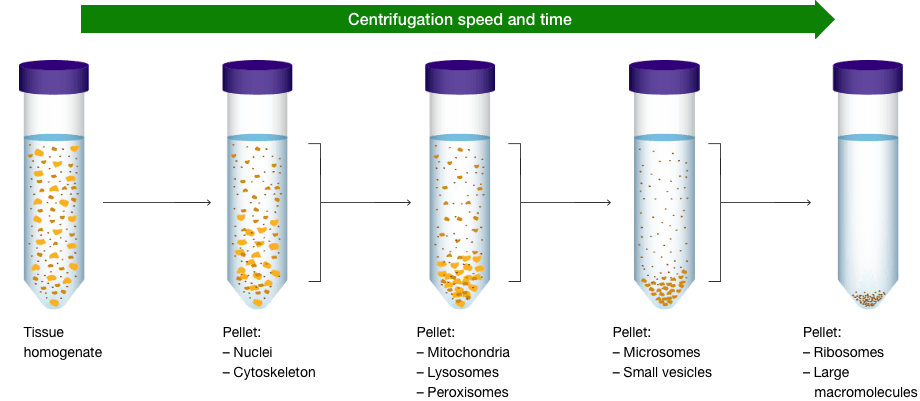
In some instances, cell organelles like mitochondria, nuclei, and endoplasmic reticulum can be separated using centrifugation following homogenization. In other instances, where enrichment of a specific organelles is necessary, cellular fractionation kits are available. This is particularly useful when probing weakly expressed proteins. The cellular fractionation process begins by first lysing cells by homogenization followed by centrifugation starting at lower speeds and gradually progressing to higher speeds. This process does require that you know where your protein of interest is expressed and the right protease and phosphatase inhibitors to use.
Protein Solubilization and Stabilization
Interactions involved in protein aggregation must be broken for successful electrophoresis. Ideally, cell lysis and protein solubilization are carried out in the electrophoresis sample buffer. If this is not possible, then proteins must be prepared in a solubilization solution that contain detergents, chaotropic/reducing agents, and buffers that will both solubilize the proteins of interest and be compatible with electrophoresis.
Lysis buffers contain different detergents that help to disrupt the cell and release proteins so that they can then be made soluble into solution. However, every protein is different and may react differently with the buffers and detergents. If you do not get your protein of interest in solution or you are studying a special protein–protein interaction, you can try different buffers and exchange the detergents.
General Guidelines
Whole-cell lysate and membrane-bound proteins — the most commonly used buffers are RIPA and NP-40. The harsh properties of RIPA buffers are best suited for hard-to-solubilize proteins.
Nuclear/mitochondria proteins — RIPA is the preferred choice here. However, fraction protocols are often first used to increase the concentration of organelle-specific target protein.
Cytoplasmic proteins — a Tris-HCl lysis sometimes shows advantages over RIPA buffer. Optimal conditions should be tested for the protein of interest.
Native protein state — CHAPS is a zwitterionic detergent that is especially well suited for protecting the native state of proteins. Commonly used for isoelectric focusing (IEF) and 2-D electrophoresis.
Nuclear/mitochondria proteins — RIPA is the preferred choice here. However, fraction protocols are often first used to increase the concentration of organelle-specific target protein.
Lysis Buffer Recipes
| NP-40 | RIPA | Tris-HCl | CHAPS |
|---|---|---|---|
| 150 mM NaCl 1% NP-40 or Triton X-100 50 mM Tris pH 8.0 |
150 mM NaCl 1% NP-40 or Triton X-100 0.5% sodium deoxycholate 0.1% SDS 50 mM Tris, pH 8.0 |
20 mM Tris-HCl, pH 7.5 | 150 mM KCl 50 mM HEPES (pH 7.4) 0.1% CHAPS |
Detergent Properties
| Detergent | Strength | Notes |
|---|---|---|
| SDS | Harsh |
|
| Triton X-100 | Mild |
|
| CHAPS | Moderate |
|
| NP-40 | Mild |
|
| Tween-20 | Mild |
|
| Urea/Guanidine | Chaotropic |
|
Reducing Agents
Many proteins exist in multimers through disulfide bonds. Applying reductants disrupts these bonds so that the proteins in the sample are present in monomeric forms. Some of the common reducing agents are below along with their typical uses.
| Reducing Agent | Properties and Applications |
|---|---|
| 2-Mercaptoethanol | Also known as beta-mercaptoethanol (BME, 2BME, 2-ME, b-mer, CAS 60-24-2), this thiol reducing agent is used to cleave protein disulfide bonds between cystine residues. |
| Dithiothreitol | Dithiothreitol (DTT, CAS 3483-12-3) is a popular protein disulfide reducing agent for sample loading buffers. |
| TCEP-HCI | Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, CAS 5961-85-3) is a strong, thiol-free reducing agent that disrupts protein and peptide disulfide bonds. |
| Cysteine-HCI | Cysteine hydrochloride (Cys-HCI, CAS 52-85-1) is used to prepare sulfhydryl assay standards or as an additive in protein refolding experiments. |
Protein Stabilization
Immediately following cell lysis, protein degradation begins to occur by proteolysis, dephosphorylation, and denaturation due to cellular enzyme activity. This activity should be kept to a minimum by preparing samples on ice or at 4˚C and by adding protease and phosphatase inhibitors to the lysis buffer, which should be freshly prepared just before use.
While there are many commercially available ready-to-use inhibitor cocktails (often proprietary), a homemade buffer blend can be made based on individual needs. The table below lists common protease and phosphatase inhibitors, their targets, and the recommended final concentration in the lysis buffer.
Common Protease Inhibitors
| Inhibitor | Protease/Phosphatase Inhibited | Final Concentration in Lysis Buffer |
|---|---|---|
| Aprotinin | Trypsin, chymotrypsin, plasmin | 2 µg/ml |
| Leupeptin | Lysosomal | 1–10 µg/ml |
| Pepstatin A | Aspartic proteases | 1 µg/ml |
| PMSF | Serine proteases | 1 mM |
| EDTA | Metalloproteases that require Mg2+ and Mn2+ | 1–5 mM |
| EGTA | Mg2+ and Mn2+ dependent proteases | 1 nM |
Common Phosphatase Inhibitors
| Inhibitor | Protease Target | Final Concentration in Lysis Buffer |
|---|---|---|
| Sodium fluoride | Serine and threonine phosphatases | 5–10 mM |
| Orthovanadate | Serine & threonine phosphatases | 1 mM |
| Pyrophosphate | Serine & threonine phosphatases | 1–2 mM |
| B-glycerophosphate | Serine & threonine phosphatases | 1–2 mM |
| Calyculin A | Serine & threonine phosphatases | 1–5 mM |
See Our Protein Extraction and Solubilization Buffers & Reagents »
Protein Sample Quantitation
It is important to know the concentration of total protein in each sample to ensure that protein gels are loaded adequately but without overloading. Additionally, proper quantitation is required to ensure different samples are compared on an equivalent basis.
The most common method to measure protein concentration is by measuring the absorbance of the lysate solution at 280 nm or 205 nm. Alternatively, several protein assays are available which rely upon the reduction of metal ions by the peptide bond, e.g., the Lowry and BCA assays, or by dye binding, as with the Bradford Assay. In all instances, a color change occurs that is proportional to the amount of protein in the sample. Protein concentration is determined by comparison of the target samples to a known standard series, typically prepared from bovine serum albumin (BSA) diluted in lysis buffer. To get the most accurate measurement of protein concentration, it is best to test a few dilutions of the sample ensuring that the results lie in the linear range of the protein assay.
Sample Loading Buffer
Once the protein concentration has been determined, samples are diluted in gel loading buffer, commonly, Laemmli sample buffer. This buffer contains glycerol, making the solution denser than the gel running buffer, so that the samples sink easily into the wells of the gel, and a tracking dye (bromophenol blue) is included, which migrates through the gel first to indicate how far the separation has progressed.
For most routine western blots, SDS (sodium dodecyl sulfate) and a reducing agent are also present in the gel loading and/or sample buffer to fully denature the protein and remove all higher-order structure. See the accompanying table for the range of loading buffer options and the supplements they contain.
Samples are heated in gel loading/sample buffer for either 5 minutes at 100°C, or 10 minutes at 70°C to aid in the denaturation. At this point, samples can remain at room temperature if they are to be used immediately or placed at 4°C or –20°C for later analysis.
Sample Buffer Conditions
| Gel Conditions | SDS | DTT or ßME | Comments |
|---|---|---|---|
| Denaturing and Reducing |
|
|
Removes all higher order structure, including disulfide bonds |
| Denaturing |
|
|
Higher order structure is disrupted, but disulfide bonds are retained |
| Reducing |
|
|
Disulfides are removed, but most higher order structure remains |
| Native |
|
|
The protein retains higher order structure. Multimers and protein-protein interactions can be detected |
Tips for Better Sample Prep
-
Prevent Degradation
After centrifuging the mixture, you will need to add lysis buffer and a protease inhibitor cocktail. You may need to repeat this process with differing concentrations of protease inhibitor cocktail to obtain a high enough protein concentration. -
Quantitate Sample
Quantitating total protein yield in your sample allows you to ensure equal loading of samples for electrophoresis. This is particularly important when needing to quantitate target intensity across lanes, for example, between control and cells undergoing experimental treatment. Equal loading minimizes the degree of data correction or normalization during analysis. -
Remove Insoluble Material before Electrophoresis
Following cell disruption, centrifuge at 20,000 x g for 15 minutes at 15°C to remove any insoluble material. Solid particles may block the pores of the gel.
Sample Preparation Protocols and Resources

Find the right products for you using the free Western Blot Selector Tool
Start Tool
Western Blotting Protocol Library
Filter by your laboratory set-up and reagents to get a custom western blotting protocol that best fits your needs.

Protein Blotting Guide
(PDF 7.1 MB)Details on blotting technology, available products, methods, tips, techniques, and troubleshooting resources.

Protein Sample Preparation
(PDF 210 KB)Protocols and tips for cell lysis, protein solubilization, and sample preparation for SDS-PAGE electrophoresis and blotting.

Fundamentals of Western Blotting Course #1: Sample Preparation

Get Your Free Protein Standard Temporary Tattoo
You too can sport a Precision Plus Protein Kaleidoscope standard tatto temporarily.
Get Yours Now
Better Western Blotting Guide
Tips, Techniques, and Technologies from the Western Blotting Experts at Bio-Rad Laboratories
Get Yours Free Guide
Western Blot University
Courses designed to make you a western blotting expert.
Enroll Today





