
On This Page |
First-Generation Sequencing | Second-Generation Sequencing | Third-Generation Sequencing | Trends in NGS | Related Topics and Products |
In 2001, the first rough draft of the approximately 3 billion nucleotides of the human genome was published as part of the international scientific collaboration known as the Human Genome Project (NHGRI 2019). This gargantuan undertaking cost nearly $1 billion and involved scientific minds from around the world who sequenced slices of the human genome in rooms full of capillary sequencers . The project was officially declared complete in 2003. Using today’s sequencing technologies, a single laboratory can complete the same project within hours and at a small fraction of the cost.
Next-generation sequencing (NGS), defined as high-throughput, massively parallel DNA sequencing, was first commercialized during the early 2000s, replacing the Sanger sequencing method that dominated the decades prior. Since then, the field has seen tremendous growth in both sequencing technologies and bioinformatics methods, going from budget-busting short reads to whole-genome sequencing at record speeds and drastically lower costs — as low as $600 per genome. With industry leaders promising NGS for $100/genome (Preston 2021), the prospects of personalized medicine will soon be within reach of every lab.
This article provides a brief history of NGS and an overview of the key technologies that characterize the different generations.
First-Generation Sequencing
In 1965, twelve years after Watson and Crick were credited with solving the three-dimensional structure of double-helix DNA from Franklin's x-ray diffraction studies showing its helical pattern, Robert Holley published the first whole nucleic acid sequence of a eukaryote - Saccharyomyces cerevisiae alanine tRNA. However, most sequencing efforts at the time were focused on microbial RNA, which had the advantage of known ribonuclease chemistries and was therefore more feasible than eukaryotic DNA. Sequencing protocols involved enzyme-fragmentation of RNA followed by laborious two-dimensional chromatography and fractionation to solve the sequence at a rate of a few nucleotides per year (Heather 2016).
The mid-1970s ushered in the “1st generation” of sequencing with Allan Maxam and Walter Gilbert’s chemical degradation, and Frederick Sanger’s dideoxy chain termination methods (Kulski 2015). These methods replaced fractionation with electrophoretic size separation on polyacrylamide gels. In Maxam–Gilbert sequencing, radiolabeled DNA is cleaved using site-specific enzymes, constituting different reaction lanes on a polyacrylamide gel. In the Sanger technique radiolabeled- or fluorescently labelled ddNTP nucleotides of each type are included in separate DNA polymerization reactions, causing chain termination at random sites of incorporation along the way. Fragments generated in both techniques are then resolved by gel electrophoresis and “read up” in order to assemble the sequence.
The First Sequencers
Sanger sequencing eventually gained dominance over Maxam–Gilbert sequencing due to its simplicity and led to the first commercially-available sequencers. These instruments simplified the fluorescent dye-terminator protocol into a single reaction and introduced computerized data acquisition and analysis with subsequent models. Developments in PCR, RT-PCR and cloning technologies enabled a flurry of published gene sequences from humans and other organisms in the late 1980s and throughout the 1990s. The first instrument for large-scale genomics applications utilized parallelized Sanger sequencing through 96 polymer-filled capillaries, yielding 9 × 105 bases per day at read lengths of up to 700 bp. This system was also the main workhorse for the Human Genome Project in 2001.
Second-Generation Sequencing
The next generation of sequencing technologies leveraged innovations in miniaturization, optical/chemical detection and nucleotide chemistry to parallelize millions of reactions in a single run, allowing deep, high-throughput sequencing with gigabases of data generated per run. The price tag for human genome sequencing saw a 50,000-fold decrease by the mid-2000s, and was approaching $1,000 by 2016. Compared to Sanger sequencing of 1st-generation platforms, NGS methods generate shorter reads and are less accurate; however, the rapid pace with which new technologies enter the market continues to challenge these limitations and make NGS more accessible to life science research and clinical applications.
Sequencing by Synthesis for NGS (Short-Read)
The basic NGS workflow includes library preparation, clonal amplification, sequencing, and analysis (Fig 1) (Goodwin 2016). During library preparation, genomic DNA is fragmented using enzymes or physical methods and ligated to platform-specific adapters. Next, template DNA is amplified via droplet or solid-state PCR in order to generate millions of template molecules for the sequencing reaction. During the sequencing reaction, each nucleotide position on the template molecule generates an optical (e.g., fluorescence) or chemical signal (e.g., pH) in response to a process, such as nucleotide addition on a growing complementary strand. This signal is recorded and analyzed computationally to determine the sequence.
-
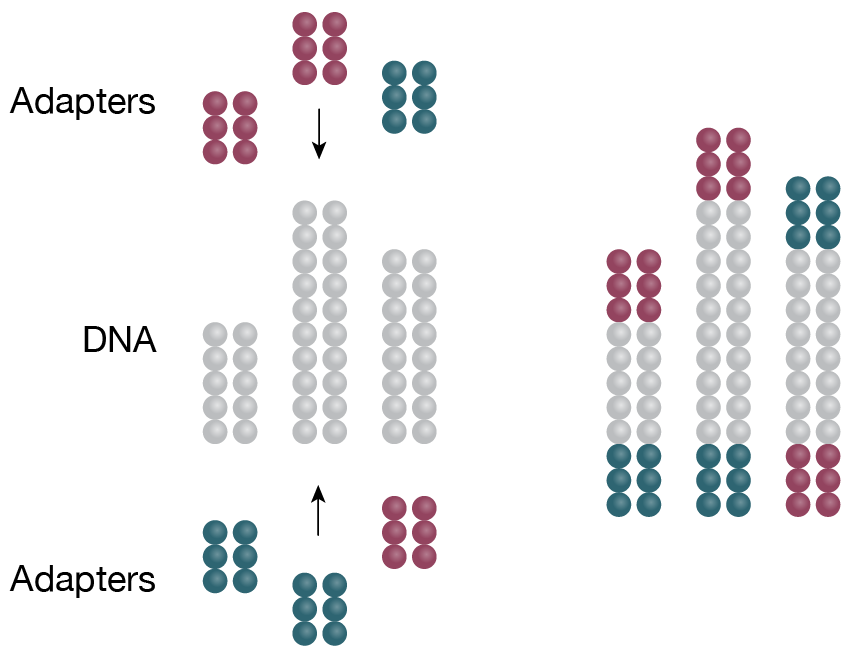
1. Library preparation
DNA is fragmented and ligated to specific adaptors in preparation for amplification -
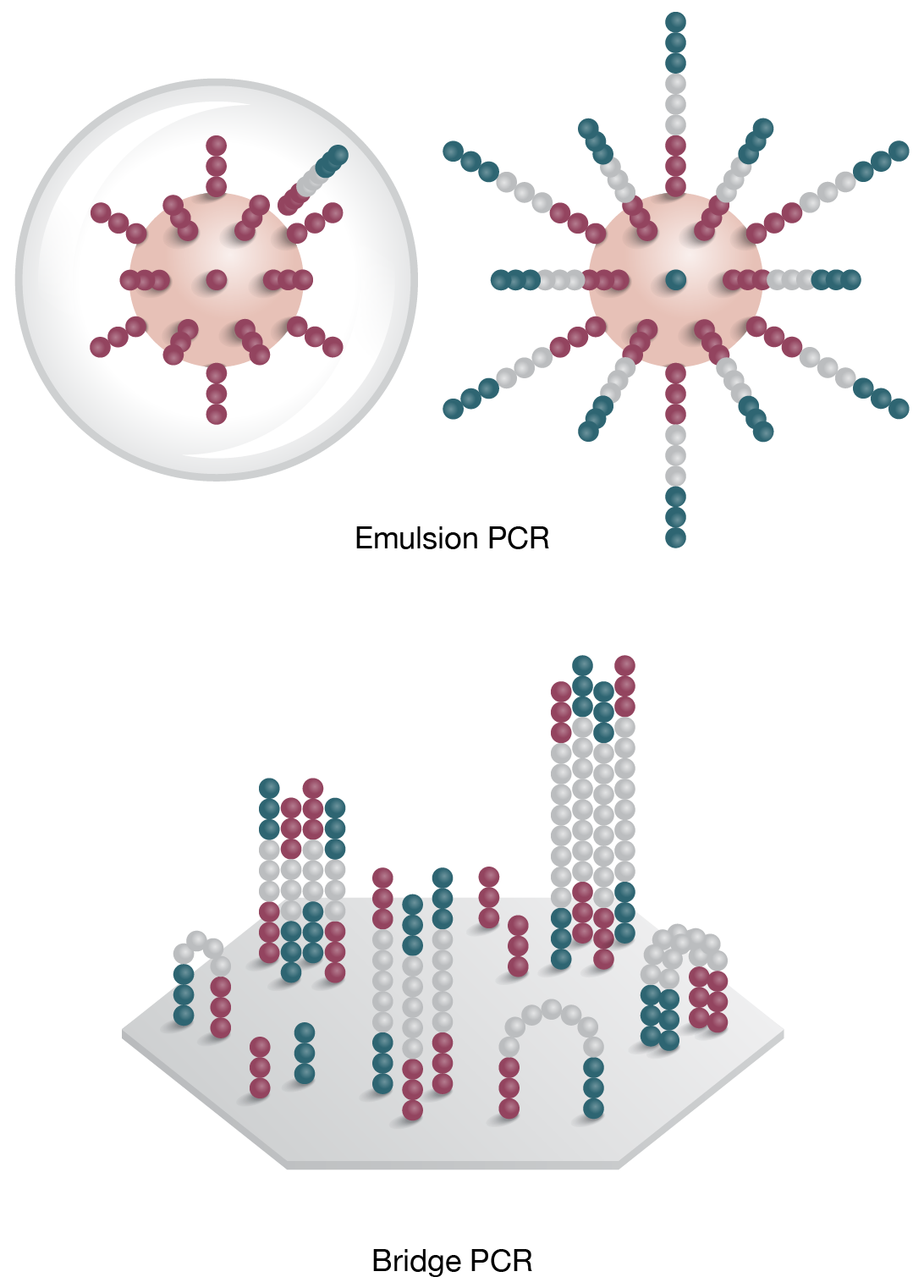
2. Clonal Ampification
Template is PCR amplified by emulsion (on bead) or bridge PCR (solid surface) -
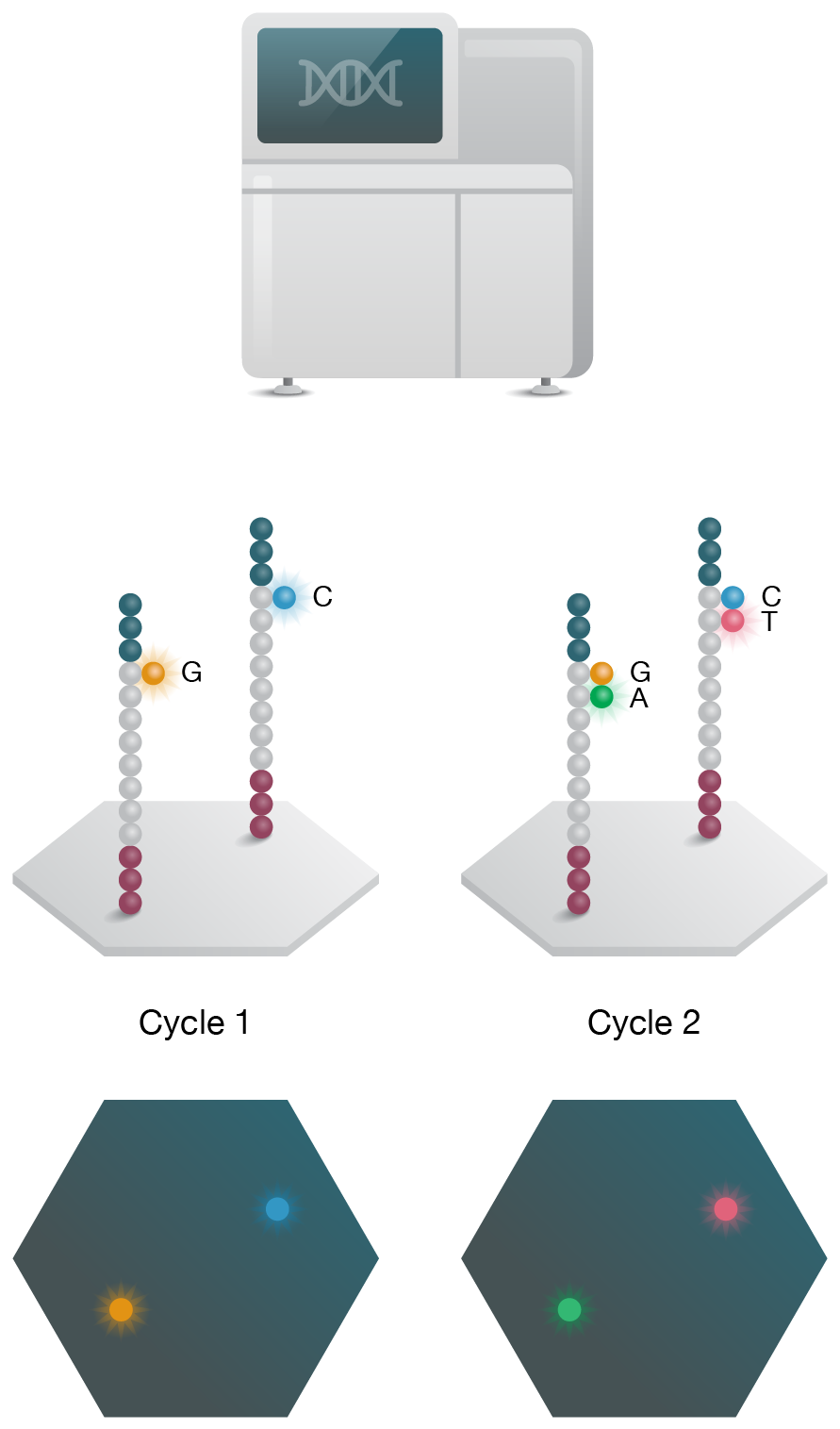
3. Sequencing
Sequencing is performed on NGS instrument and the output signal is recorded -
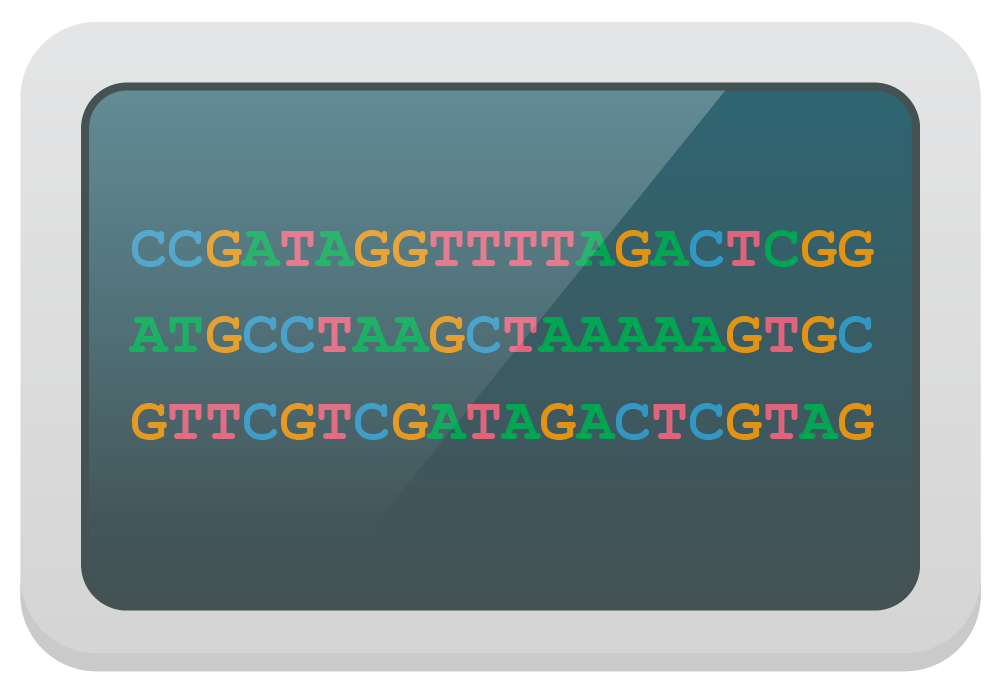
4. Data Analysis
Optical or chemical signal is processed to determine sequence
Figure 1: Basic NGS workflow.
Amplification formats for Emulsion PCR versus Bridge PCR
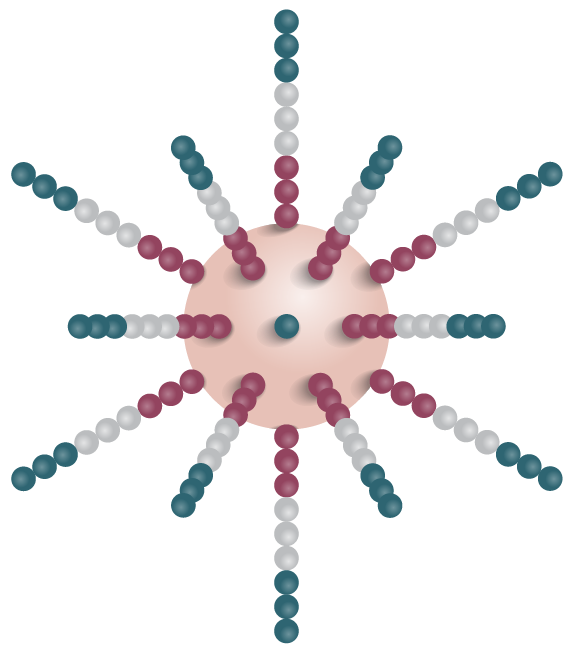
Emulsion PCR and bridge amplification are the two predominant methods used to generate millions of copies of template, or PCR colonies (polonies). In the emulsion PCR method, adapter-ligated template, primers, polymerase and beads (displaying complementary adapters) are packed inside micelles, where on-bead PCR generates thousands of PCR clones (Fig 2A). In solid-phased amplification “bridge PCR” is used to create clone clusters on the surface of a flow cell, which is then accessible to the sequencing reaction (Fig 2B).
A. Emulsion PCR - used in pyrosequencing, proton sequencing, and sequencing-by-ligation (SBL)
-

1. Adapter-ligated template, primers, polymerase and beads are contained within micelles.
-
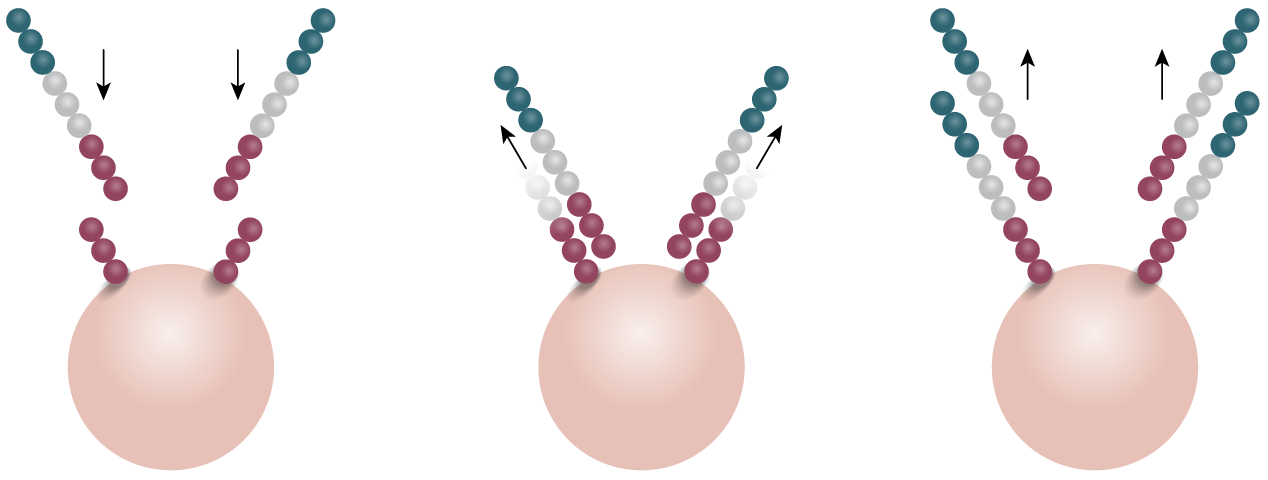
2. Template binds complementary primers on the bead. PCR generates the complementary strand.
-
3. Beads with clonal polonies are then used in sequencing reaction.
B. Bridge PCR - used in industry-leading short-read,sequencing-by-synthesis (SBS)

1. Adapter-ligated template binds complementary primer on surface.
2. Free ends of template bind complementary primer forming a "bridge".

3. Bridge PCR generates the complementary strand.

4. Strands denature and form new bridges with free primers.
5. Process repeats to form clone clusters.
Figure 2: Emulsion and bridge PCR. A) In emulsion PCR, clonal amplification takes place inside an emulsion droplet, producing beads that are bound to thousands of template molecules. B) In solid-state bridge PCR, repeated cycles of template amplification on a lawn of primers generates clone clusters.
Similar to Sanger sequencing, the first NGS technologies were sequencing by synthesis (SBS) protocols that require polymerase chain extension. However, instead of labeled dNTPs and physical separation of fragments, a two-step chemistry is used to measure the resulting chemiluminescence upon pyrophosphate release at each dNTP incorporation step. Since the signal produced in pyrosequencing does not distinguish between different nucleotides, each of four nucleotides must be introduced in turn.
Pyrosequencing was featured in the first NGS instrument to hit the market in 2005. This system distributes template-bound beads in a picotiter plate and uses a charge-coupled device camera to record the chemiluminescence signal during chain elongation. The first NGS instrument series produce superior read lengths of up to 700 bp, but suffer due to long run times (> 10 hr) and low accuracy within homopolymer regions. Pyrosequencing has been largely replaced by newer technologies.
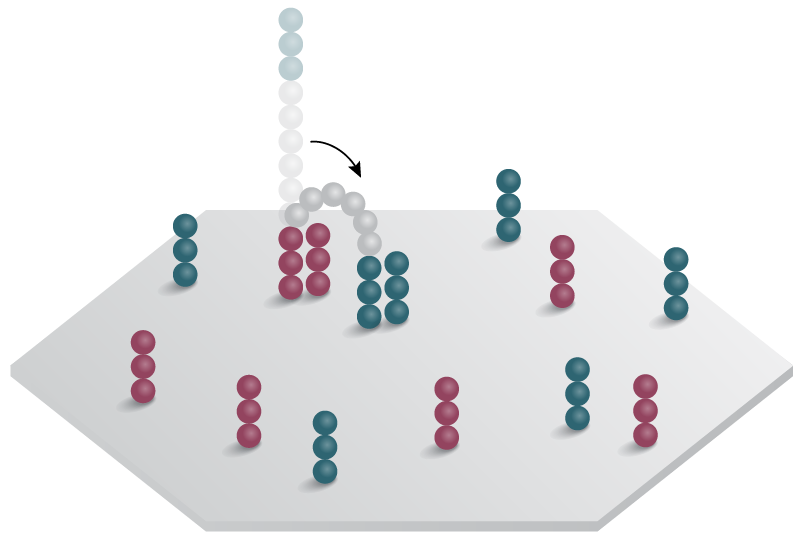
The most popular SBS method is a dye-sequencing technology that dominates the short-read NGS market with a suite of instruments from low-throughput (0.3–15 Gb) benchtop instruments to ultra-high-throughput (120–1,500 Gb) systems for large-scale genomic studies (Fig 3A) (Goodwin 2016). Unlike the first pyrosequencing instruments, this method uses a cocktail of removable fluorescently labeled chain-terminating dNTPs with four unique colors. This distinction also yields more accuracy within homopolymer regions. Amplification of adapter-ligated template is achieved using PCR “bridge amplification” on a solid surface to generate clusters of clones. During DNA polymerization, the surface is imaged at each dNTP incorporation step, before un-blocking the chain for the next cycle. This technology offers competitive cost/bp, read lengths (up to 300 bp) and accuracy due to built-in paired-end sequencing; however, the runtimes (hours to days) are not amongst the fastest on the market.
Sequencing by Ligation for NGS (Short-Read)
While SBS methods rely on the action of DNA polymerase, sequencing by ligation (SBL) approaches use DNA ligase to extend the complementary strand (Goodwin 2016). The four nucleotide probes are introduced in sequence, and the reaction is imaged after each reaction cycle. One popular SBL method uses hybridization/ligation of dinucleotide-labeled probes in a four-color detection system. Given the 16 possible combinations of dinucleotides, each color is associated with four unique dinucleotide pairs (e.g., red = AT, CG, GC, and TA). As each color signal can be associated with one of four different dinucleotides, the final sequence is determined using bioinformatics analysis. Further, the dinucleotide probes introduce exogenous anchor nucleotides into the growing strand, producing gaps in data. Full template coverage is achieved with rounds of repetition that are designed with single-nucleotide offsets, allowing dinucleotide ligation at positions occupied by anchor nucleotides in previous rounds. These technologies afford excellent accuracy (~99.99%) and low cost, but are limited by very short read lengths and long run times.
Beyond Light: Proton Sequencing
The group that invented the first pyrosequencing instruments developed and commercialized ion semiconductor sequencing (also known as proton sequencing) in 2010 (Goodwin 2016). The fundamental difference between proton sequencing and other SBS sequencing technologies is the lack of optical detection during strand elongation. Instead, ion sensors beneath the flow cells detect when a nucleotide is incorporated into a growing DNA chain, because this addition releases a hydrogen ion and causes a pH change in the solution. When a specific dNTP is introduced, the release of hydrogen ions can indicate if zero, one or more nucleotides were incorporated. If an A, T, G or C nucleotide does not lead to hydrogen release, then it is not complementary to the sequence. The shift in pH is not perfectly correlated with the number of incorporation events; thus, proton sequencing is less accurate in homopolymeric stretches and can lead to premature sequence truncation. Compared to other technologies, proton sequencing can offer longer read lengths (up to 400 bp), and short run times (as low as 2 hours) at a competitive price point. It is commonly used for amplicon sequencing or smaller targeted regions within genomes.
Third-Generation Sequencing
While standard short-read sequencing technologies dominate the current NGS market, newer third-generation instruments have opened the door to long-read, direct-read, and single-cell sequencing (Goodwin 2016; Stark 2019). These technologies address some of the key limitations of short-read sequencing and are becoming more prevalent in the field of medical genetics.
Collectively, third-generation sequencing technologies eliminate some of the defining features of other generations, while introducing new capabilities with respect to single-molecule and real-time sequencing, enabling continuous long reads of up to 200 kb (Kulski 2016). These technologies allow scientists to better interrogate the complexities of long-range genomic structures, at considerately lower costs.
-
A. Sequencing by synthesis (Short Read)

-
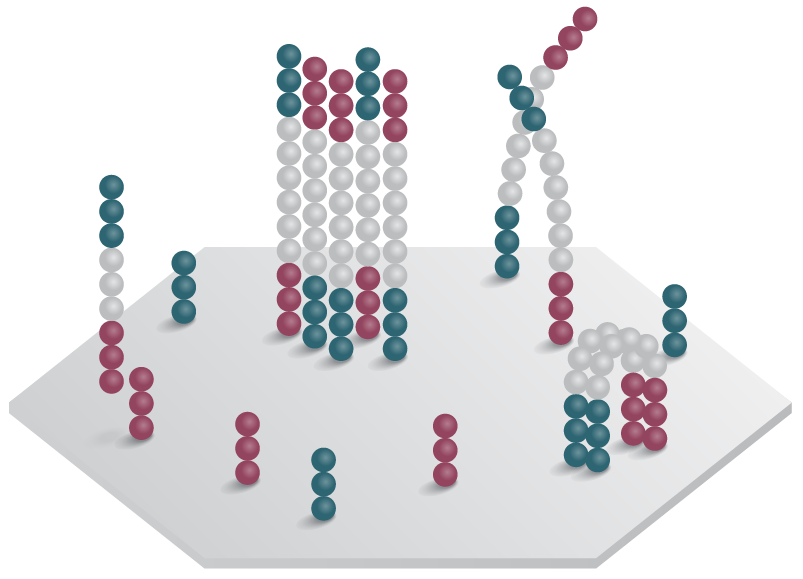
B. Single-molecule real-time (Long Read)
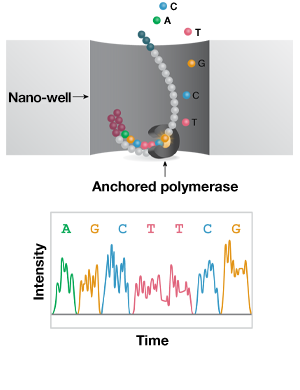
-
C. Nanopore (Direct Long-Read)

Figure 3: Short- and long-read NGS platforms. A) In the industry-leading short-read NGS platform, template DNA is amplified by bridge PCR to generate clusters on a flowcell. In each reaction cycle, fluorescent nucleotides are introduced, generating a color signal when incorporated into the growing strand. B) In single-molecule real-time (SMRT) long-read sequencing, individual molecules of adapter-ligated template are bound to a single polymerase molecule that is fixed to the bottom of a nanowell. During DNA polymerization of the complementary strand, the wells are imaged to identify the incorporated fluorescent nucleotide. C) In nanopore single-molecule long-read sequencing, the template is ligated to a motor protein and delivered to a flowcell. Here, the motor protein threads the template through a nanopore, where a change in current is associated with specific nucleotides.
Long-Read Sequencing
In contrast to short-read protocols that read a few hundred nucleotides at a time, long-read sequencing technologies can read up to 200 kb of DNA in a continuous fashion. This difference in read length is significant when studying genomic regions with high GC content, repetition, and structural variations. These regions pose a challenge when assembling sequences from short reads, even with the most sophisticated computational algorithms. Importantly, the lack of a PCR step in contemporary protocols reduces coverage bias of GC-heavy regions, some of which may be implicated in human diseases.
The key technologies in this space are single-molecule real-time (SMRT) sequencing and nanopore sequencing.
In SMRT sequencing, a form of sequencing-by-synthesis reaction takes place at the bottom of nanowells, where individual polymerase molecules process one molecule of a template, per well. Nanopore sequencing utilizes a helicase-pore complex to couple each nucleotide to a defined shift in electrical current as it passes through; this allows “direct” sequencing of the DNA or RNA template. These “physical” approaches differ from synthetic long reads that computationally reconstruct sequences from short-read data collected on barcoded fragments.
Long-read sequencing is a powerful tool for genomics and transcriptomics studies and a complement to existing short-read methodologies (Mantere 2019). These technologies have been used to resolve allele phasing, discriminate pseudogenes, map structural variants, and sequence tandem repeat regions, all of which can be important in the etiology of human diseases, as shown in a recent allele phasing study involving cystic fibrosis patients (Regan 2015). In cancer research, long-read sequencing is capable of genotyping complicated cancer genomes, including structural variants and short indels. Improvements in accuracy, cost, and data analysis will further integrate long-read sequencing in diagnosis, treatment planning, and other facets of clinical care.
Single-molecule long-read sequencing
One key difference between single-molecule, long-read sequencing technologies and short-read protocols is the lack of a clonal amplification step. The single-molecule real-time (SMRT) long-read sequencing platform is currently the most popular on the market (Fig 3B). In this system, strand elongation takes place in zero-mode waveguides (ZMWs), which are arrays of microfabricated nanowells with transparent bottoms. One polymerase molecule is fixed to the bottom of each well, such that the physical location of nucleotide incorporation within the well remains constant. During polymerization, each dNTP incorporation is imaged just before the fluorescent probe is cleaved, resulting in a unique color signal for each dNTP. SMRT sequencing allows fast, real-time sequencing of long templates (up to 20 Kb), and accurate short reads via repeated sequencing of a circularized template.
Nanopore sequencing, first commercialized in 2014, is a single-molecule system for direct sequencing of native DNA and RNA with long-read potential of 200 kb (Fig 3C). Unlike SMRT and other sequencing methods, nanopore sequencing does not measure optical or chemical signals from polymerization cycles of a complementary strand. Instead, it uses the action of a motor protein to denature dsDNA and thread the resulting ssDNA through a protein channel that is filled with an electrical current. Depending on the structure, each nucleotide causes a unique shift in voltage as it passes through the pore, which is then analyzed to solve the sequence. Nanopore sequencing has been associated with high-error rates for ultra-long and repeat regions, but newer protocols continue to address these limitations. Chips capable of running a few thousand pores on 48 flow cells place this technology on par with high-throughput second-generation instruments.
Synthetic long-read sequencing
Synthetic sequencing leverages available short-read technologies to construct long molecule sequences computationally (Goodwin 2016). Long-template DNA is fragmented and barcoded inside microtiter plate wells or emulsion drops, followed by short-read sequencing on existing devices. The sequence data is then sorted in silico and reassembled into long strands according to the barcode identifiers. The two current systems for synthetic long-read sequencing deliver yields up to 100 kb of synthetic length (speed depends short-read technology used).
Trends in Next-Generation Sequencing Technologies
The rapid advancement of next-generation sequencing (NGS) technologies over the last two decades has revolutionized the field of genomics and made sequencing accessible to many research and clinical applications such as transcriptomics and gene expression, genome-typing, and epigenomic studies of many different organisms (Goodwin 2016; Stark 2019). Established and new market players are now focused on pushing the limits of speed and cost, helping establish sequencing as a routine part of clinical care. Moreover, newer generations of instruments continue to make gains in areas of long range, direct, and single-cell sequencing, in addition to introducing novel types of measurement. Here we highlight a handful of technologies and applications that are at the forefront of these efforts, primarily in the area of long-range sequencing.
Spatially Resolved Sequencing
While standard bulk and single-cell sequencing data is disconnected from cellular context, spatialomics preserves spatial information and allows for a better understanding of complex diseases (Burgess 2019). As an example, in a recent study, gene expression profiles were mapped across prostate cancer tissue samples, revealing “high-risk” cancer-prone areas beyond the annotated tumor boundaries (Berglund 2018). In transcriptomics studies, spatial encoding and in situ transcriptomics capture spatial data prior to sequencing using either laser capture micro-dissection or barcoding of RNAs prior to isolation. These methods have been successfully used on tissue sections from a variety of species and report abundances with more sensitivity than traditional fluorescence in situ hybridization (FISH)-based methods. The most recent technology, high-throughput fluorescent in situ sequencing method (FISSEQ) allows in situ RNA sequencing directly within tissue, cell culture, and whole-mount embryos.
Point-of-Care Sequencing
NGS is an invaluable clinical tool for identifying genetic factors that cause disease or determine response to a given treatment (Pereira 2017). Multi-gene panels are being considered as a cost-effective approach for identifying the genetic basis of complex diseases and cancers of unknown origin. Whole-genome and whole-exome sequencing can potentially reveal rare genetic disorders and cancer risk factors, but due to their high costs and potential for discoveries without clear clinical meaning, these methods are employed on a case-by-case basis. Clinical oncologists can now use RNA sequencing for cancer typing and profiling tumor and immune checkpoint markers to assess treatment response, while newer protocols are being applied to circulating tumor DNA and RNA (ctDNA and ctRNA) discovery. While some diagnostic genetic tests (e.g., prenatal testing) are already approved for use, wide adoption of genetic testing in the clinic faces many challenges. Data reliability, data interpretation, quality control, and best practices for communicating results with patients are among the important questions that must be addressed.
Rare Disease Diagnosis
The diagnosis of rare genetic diseases (defined as < 1/15,000 in the US) has historically been a long, often watch-and-wait process, with late access to sequencing information (Liu 2019). NGS has dramatically changed this workflow by allowing genomic testing early on, providing significant benefits to patients. Over the last decade, whole-exome and genome sequencing have helped identify disease-causing genes in rare metabolic, neurodevelopmental, neuromuscular, movement, and demyelinating disorders (Fernandez-Marmiesse 2018). Of great interest are gene panels that focus on the coding regions of relevant genes as a cost-effective and practical NGS-based diagnostic tool for rare diseases. As with most clinical tools, accuracy and data interpretation is an ongoing challenge. Additionally, there are ethical questions around responsible handling of data and communication to patients, in particular with direct-to-consumer test kits.
Infectious Disease Sequencing
NGS is very applicable and versatile for pathogen characterization. Short-read sequencing platforms for routine sequencing of up to 1000 basepair fragments are common in most clinical and public health microbiology laboratories in the US. Long-read sequencing platforms such as single-molecule real-time sequencers can construct complete genomes very accurately (Gwinn M, 2019). More recently, epidemiologic studies in the US and Europe have capitalized on the ability of NGS to genotype the full mutation profile specific to a given SARS-CoV-2 variant (Wurtz N, 2021).
Drug Discovery & Immunotherapy
Genome Editing (CRISPR/Cas9)
CRISPR/Cas9 is a powerful genome editing tool that allows precise changes to the DNA of living cells and animals. These methods have seen expanded use in the research setting, with the promise of a cure for previously untreatable genetic disorders. However, the off-target activity of these methods makes them far too risky for human therapeutics. NGS methods are widely used to detect off-target effects in vitro or in vivo (Vakulskas 2019; Zhang 2015). In vitro, digested genome sequencing methods detect double-strand breaks in cell-free genomic DNA, while in vivo methods identify double-strand breaks and associated rearrangements in different cell types under varied conditions. Data analysis remains a significant obstacle, as there is a current lack of adequate commercial solutions.
Immune Repertoire Sequencing
Cancer immunotherapies, including checkpoint inhibitors, adoptive-cell therapies, and therapeutic vaccines have demonstrated immense success in patients. However, drug resistance and limited efficacy in many patient populations necessitates a better understanding of the molecular mechanisms involved. Studies of the human immune repertoire, which is the collection of diverse T cell and B cell receptors, are used to better dissect immunotherapy responses, with the aim of designing personalized treatments to serve broader patient populations (Zhuang 2019). Immune repertoire sequencing (IRS) of DNA and RNA is favored over proteomics methods for large-scale analysis of the immune repertoire and is combined with other technologies to identify new vaccine targets. Wide application of IRS in basic science and clinical oncology hinges on advancements in existing bioinformatics tools and protocols.
Conclusion
The future of genomics belongs to rapidly evolving third-generation sequencing methods (long-read, direct-read, and single-cell) and new technologies around in situ sequencing and spatialomics. As new technologies emerge at an unprecedented rate, so do questions and challenges around data storage, quality control, regulatory guidelines, and the ethics of handling such data. Further, reduced costs and better bioinformatics approaches will help close the translation gap, and provide physicians with genomics tools for delivering timely, personalized care.
Given the sheer number of technical innovations in genome sequencing, it is hard to believe the field of NGS is only a few decades old. The genomics revolution, which brought the Human Genome Project to fruition in the early 2000s, has since produced population-level genomic insight through the 1000 Genomes Project (2010) and other projects alike, and made whole genome sequencing accessible to clinical applications. The rapidly expanding applications of NGS strategies to transcriptomics, epigenomics, and other –omics fields continue to shed light on the relationship between genomics and human disease. As new sequencing technologies like nanopore sequencing push the limits of speed, accuracy, read length and cost, NGS gains more relevance not only as a powerful research tool but also as an indispensable part of medical practice.
Related Topics and Products

PCR (Polymerase Chain Reaction)
Learn how PCR works, how to choose the right PCR instrument, how to design, optimize, analyze, and troubleshoot PCR assays.

Real-Time PCR (qPCR)
Discover an array of solutions to optimize, execute, and troubleshoot MIQE-compliant real-time PCR experiments.

Introduction to Digital PCR (ddPCR)
Learn how breakthrough digital PCR technology is used to provide ultrasensitive and absolute nucleic acid quantitation.

Advances in RNAseq
NGS RNA sequencing (RNA-Seq) is the foremost tool for transcriptomics, gene expression profiling, and the study of differential RNA splicing.

RNA-Seq Workflow
A guide to the steps of an RNA-Seq workflow including library prep and quantitation and software tools for RNA-Seq data analysis.
The Non-Coding Transcriptome
Explore the diverse classes of noncoding RNA, the origins of ncRNA species, their biological roles, and clinical implications.
SARS-CoV-2 / COVID-19 Assay and Research Solutions
Bio-Rad provides a wide range of products for use in the support of COVID-19 diagnostic screening, confirmation of test results, surveillance, and therapeutic & vaccine research and development.

ddPCR Master Class Videos
Become an expert in all things ddPCR. Our master class sessions allow you to learn from the best in the field. From instrument overviews to informational sessions highlighting the implications of ddPCR in the real world, Master Classes have it all.

Targeted Genome Editing
Learn about the major systems for targeted genome editing, including CRISPR/Cas9, some key applications, and workflow options.

Traditional PCR Systems
DNA amplification instruments range from personal thermal cyclers to the flexible 1000-series.

qPCR Detection Systems
These systems deliver sensitive, reliable detection of both singleplex and multiplex real-time PCR reactions.

Digital PCR
Bio-Rad's unique Droplet Digital PCR technology provides absolute quantification of nucleic acids for a wide range of applications including cancer mutation studies, HIV quantification, and environmental monitoring.

PCR Plastic Consumables
A large selection of thin-wall PCR tubes, plates, seals, and accessories precisely manufactured for optimal fit and cycling performance in Bio-Rad thermal cyclers, real-time PCR systems, ddPCR systems, and all major competitor systems.

PCR & qPCR Reagents
A wide range of reagents for reverse transcription, PCR, and real-time PCR, optimized to generate accurate and reproducible data.
Next-Generation Sequencing
Novel products for NGS library preparation and quantitation used in cutting-edge applications such as whole-transcriptome RNA-Seq and single-cell sequencing.

PrimePCR PCR Primers, Assays & Arrays
Experimentally validated PCR primer and probe assays for gene expression, copy number variation, and mutation detection analysis for real-time PCR and Droplet Digital PCR.
Droplet Digital PCR Assays
Bio-Rad offers a comprehensive portfolio of Digital PCR Assays and Kits for numerous applications including Mutation Detection, Copy Number Determination, Genome Edit Detection, Gene Expression, Expert Design Assays, Residual DNA Quantification and Library Quantification.
References
Barros-Silva D, Marques CJ, Henrique R, Jerónimo C. Profiling DNA Methylation Based on Next-Generation Sequencing Approaches: New Insights and Clinical Applications. Genes (Basel). 2018 Aug; 9(9):429. PMCID: PMC6162482.
Berglund E, Maaskola J, Schultz N, et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun. 2018; 9(1):2419. PMCID: PMC6010471.
Bik EM. The Hoops, Hopes, and Hypes of Human Microbiome Research. Yale J Biol Med. 2016 Sep; 89(3):363-373. PMCID: PMC5045145.
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015 Jan; 109:21.29.1-21.29.9. PMCID: PMC4374986.
Burgess DJ. Spatial transcriptomics coming of age. Nat Rev Genet. 2019 Jun; 20(6):317. PMID: 30980030.
Cheng M, Cao L, Ning K. Microbiome Big-Data Mining and Applications Using Single-Cell Technologies and Metagenomics Approaches Toward Precision Medicine. Front Genet. 2019 Oct; 10:972. PMCID: PMC6794611.
Ciccarone F, Tagliatesta S, Caiafa P, Zampieri M. DNA methylation dynamics in aging: how far are we from understanding the mechanisms? Mech Ageing Dev. 2018 Sep; 174:3-17. PMID: 29268958.
Fernandez-Marmiesse A, Gouveia S, Couce ML. NGS Technologies as a Turning Point in Rare Disease Research , Diagnosis and Treatment. Curr Med Chem. 2018 Jan; 25(3):404-432. PMCID: PMC5815091.
Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016; 17(6):333-51. PMID: 27184599.
Gwinn M, MacCannell D, Armstrong GL. Next-Generation Sequencing of Infectious Pathogens. JAMA. 2019;321(9):893–894. doi:10.1001/jama.2018.21669.
Human Cell Atlas: https://www.humancellatlas.org/.
Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018; 50(8):96. PMCID: PMC6082860.
Jin Z, Liu Y. DNA methylation in human diseases. Genes Dis. 2018;5(1):1–8. PMCID: PMC6147084.
Liu Z, Zhu L, Roberts R, Tong W.Toward Clinical Implementation of Next-Generation Sequencing-Based Genetic Testing in Rare Diseases: Where Are We? Trends Genet. 2019 Nov; 35(11):852-867. PMID: 31623871.
Mantere T, Kersten S, Hoischen A. Long-Read Sequencing Emerging in Medical Genetics. Front Genet. 2019; 10:426. PMCID: PMC6514244.
Pereira M, Malta F, Freire M, and Couto P. Application of next-generation sequencing in the era of precision medicine. In: Marchi F, Cirillo P, Mateo EC (Eds.) Applications of RNA-Seq and Omics Strategies: From Microorganisms to Human Health. InTechOpen, London, England, UK; 2017: 293–318. doi: 10.5772/intechopen.69337.
Preston J, VanZeeland A, and Peiffer DA. Innovation at Illumina: The road to the $600 human genome. Nature Portfolio Feb 10 2021.
Regan JF, Kamitaki N, Legler T, et al. A rapid molecular approach for chromosomal phasing. PLoS One. 2015; 10(3). PMCID: PMC4349636.
Sarda S, Hannenhalli S. Next-generation sequencing and epigenomics research: a hammer in search of nails. Genomics Inform. 2014 Mar; 12(1):2-11. PMCID: PMC3990762.
Shinde P, Mohan L, Kumar A, et al. Current Trends of Microfluidic Single-Cell Technologies. Int J Mol Sci. 2018; 19(10):3143. doi: 10.3390/ijms19103143.
Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019; 20(11):631-656. PMID: 31341269.
Vakulskas CA, Behlke MA. Evaluation and Reduction of CRISPR Off-Target Cleavage Events. Nucleic Acid Ther. 2019 Aug; 29(4):167-174. PMCID: PMC6686686.
Wurtz N, Revol O, Jardot P, Giraud-Gatineau A, Houhamdi L, Soumagnac C, Annessi A, Lacoste A, Colson P, Aherfi S, Scola B. Monitoring the Circulation of SARS-CoV-2 Variants by Genomic Analysis of Wastewater in Marseille, South-East France. Pathogens. 2021 Aug 17;10(8):1042. doi: 10.3390/pathogens10081042. PMID: 34451505; PMCID: PMC8401729.
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015 Nov ; 4(11):e264. PMCID: PMC4877446.
Zhuang Y, Zhang C, Wu Q, Zhang J, Ye Z, Qian Q. Application of immune repertoire sequencing in cancer immunotherapy. Int Immunopharmacol. 2019 Sep; 74:105688. PMID: 31276974.
