
On This Page |
Types of Noncoding RNAs | Biological Roles of Noncoding RNAs | ncRNAs as Research Tools | Diagnostic Applications of ncRNAs | Therapeutic Applications of ncRNAs | Related Topics and Products |
Beyond the central dogma of molecular biology lies the noncoding transcriptome, in which, unlike messenger RNA (mRNA), transcribed RNA is not translated into protein. Such noncoding RNAs (ncRNAs) comprise over 80% of the transcriptome by mass and are linked to a growing list of functional roles in human biology and disease (Pallazo 2015).
Housekeeping ncRNAs, ribosomal RNA (rRNA) and transfer RNA (tRNA), were discovered and extensively studied in the 1950s. Other ncRNAs, starting with small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA) were not identified until the 1980s. Microarray technology and, later, the advent of next-generation sequencing (NGS) in the early 2000s greatly broadened our understanding of ncRNAs and introduced new opportunities for clinical applications. This article defines the different classes of ncRNA and highlights some of the known functional and clinical implications.
Types of Noncoding RNAs
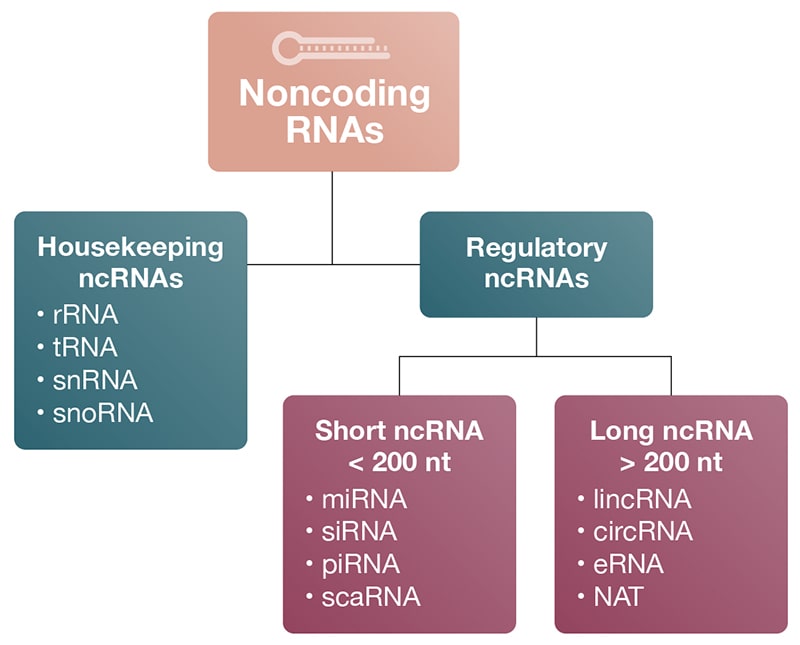
While ncRNAs are related in that they are not translated into proteins, they differ considerably in their biogenesis, size, and function (Delpu 2016). All ncRNAs are classified based on nucleotide length as either small ncRNAs (< 200 nt), or long ncRNAs (> 200 nt) (Fig 1; Table 1). The 200-nucleotide cutoff is an arbitrary designation so exceptions to the rule do exist.
Figure 1
-
A
 B
B
Figure 1. Types and abundances of ncRNAs. A) Classification of ncRNAs: Noncoding RNAs are classified into housekeeping and regulatory RNAs based on their respective roles in biology. Regulatory ncRNAs are further divided into short and long ncRNAs based on their nucleotide length. Housekeeping RNAs, snRNA, and snoRNA are also considered short RNAs because they have fewer than 200 nucleotides. B) ncRNA abundances by mass (Pallazo 2015).
| Type | Length (nt) | Function | |
|---|---|---|---|
| Small ncRNAs (<200 nt) | MicroRNA (miRNA) | 18–22 | RNAi; Protein translation regulation |
| Small interfering RNA (siRNA) | 20–25 | RNAi; Antiviral defense | |
| Piwi RNA (piRNA) | 26–31 | Regulation of transposable elements | |
| Trans-activating CRISPR (tracr) RNA | ~65 | CRISPR/Cas adaptive immunity in bacteria | |
| Small nuclear RNA (snRNA) | 100–300 | Intron splicing; RNA processing | |
| U-rich snRNA (snRNA) | 100–300 | snRNA subclass; intron splicing | |
| Small nucleolar RNA (snoRNA) | 60–200 | rRNA processing | |
| Signal recognition particle RNA (7SL) | 300 | RNA component of the signal recognition particle (SRP); protein synthesis | |
| Y RNAs | 80–120 | Components of ribonucleoproteins; DNA replication and RNA processing | |
| Small Cajal body-specific RNA (scaRNA) | 200–300 | Biogenesis of small nuclear ribonucleoproteins | |
| Long ncRNAs (>200 nt) | Long intergenic ncRNA (lincRNA) | 1 kb | Protein scaffolding |
| Natural antisense transcript (NAT) | >200 | RNAi, alternate splicing, genome imprinting | |
| Circular RNA (circRNA) | 100–999 | miRNA decoy, protein regulation | |
| Housekeeping ncRNAs | Ribosomal RNA (rRNA) | >1,500 | Protein synthesis |
| Transfer RNA (tRNA) | 76–90 | Protein synthesis | |
Table 1: Examples of ncRNAs, average nucleotide (nt) lengths and known function(s) (Pallazo 2015; Kowalski 2015).
Housekeeping ncRNAs, rRNA, and tRNA perform essential roles in protein translation (Lodish 2000; Bhagavan 2011). Ribosomal RNA is the most abundant RNA in the cell, and together with ribosomal proteins, forms the ribosomes, which slide along the mRNA and catalyze the assembly of a polypeptide chain from amino acids.
tRNAs (Fig 2A) carry unique codons that essentially decode the mRNA transcript and recruit the correct amino acids to an elongating polypeptide chain. Catalytic housekeeping RNAs called ribozymes are involved in the biogenesis of mature tRNA; ribonuclease P is a ribozyme that processes pre-tRNA into mature tRNA via its catalytic RNA component. Mature tRNA is also processed into small fragments called tRNA-derived fragments. These tRNA fragments are highly conserved and can act to regulate gene expression via the RNA interference (RNAi) pathway or inhibit protein translation (Keam 2015).
Small ncRNAs are characterized by lengths shorter that 200 nt (Delpu 2016). In this category, single-stranded micro RNAs (miRNAs) (Fig 2B) are the most thoroughly studied, with over 38,589 sequences deposited in the miRBase (Release 22.1) database as of January 2022 (www.mirbase.org). The biogenesis process in eukaryotic cells begins when DNA is transcribed by RNA polymerase II, followed by various steps that include the action of the Dicer enzyme, an endoribonuclease that cleaves double-stranded pre-miRNA into short double-stranded miRNA fragments. Mature miRNAs act via an RNA-induced silencing complex (RISC) to target and neutralize specific mRNAs as part of the RNAi pathway. Exogenous double-stranded small interfering RNAs (siRNAs), another Dicer product, use similar machinery to the RNAi pathway; however, unlike miRNAs, they target mRNAs more specifically. Mature siRNAs resemble miRNAs but are processed from longer double-stranded RNA, whereas siRNAs are derived from shorter RNA hairpin-loop structures.
Figure 2
-
A
 B
B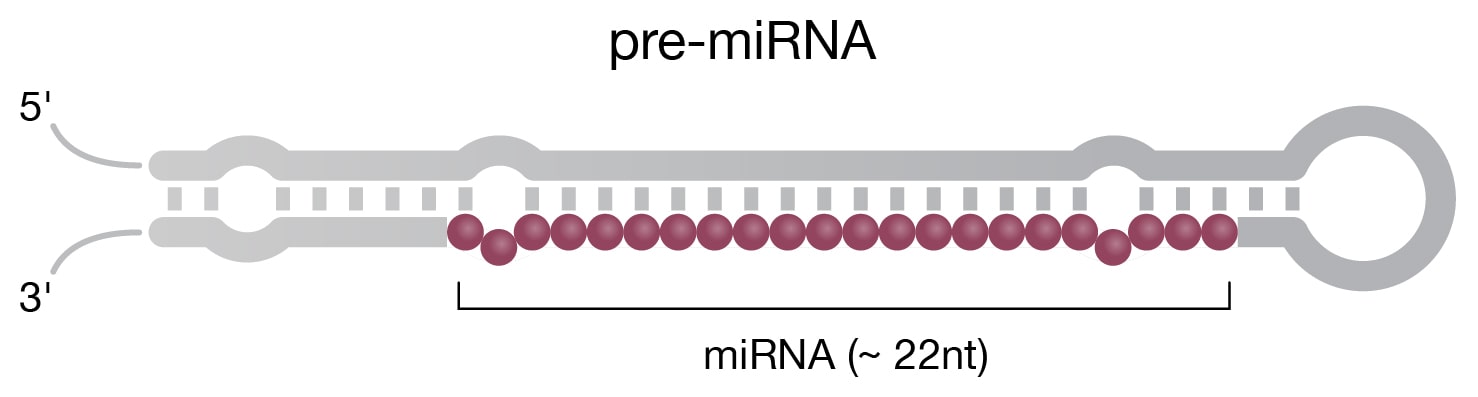
-
C
 D
D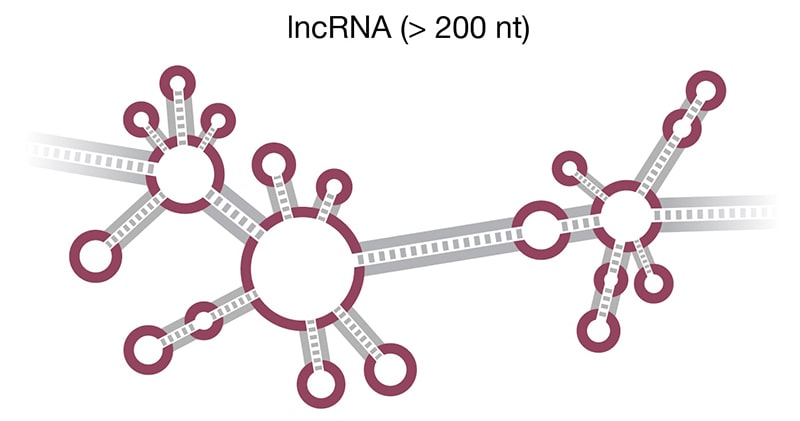
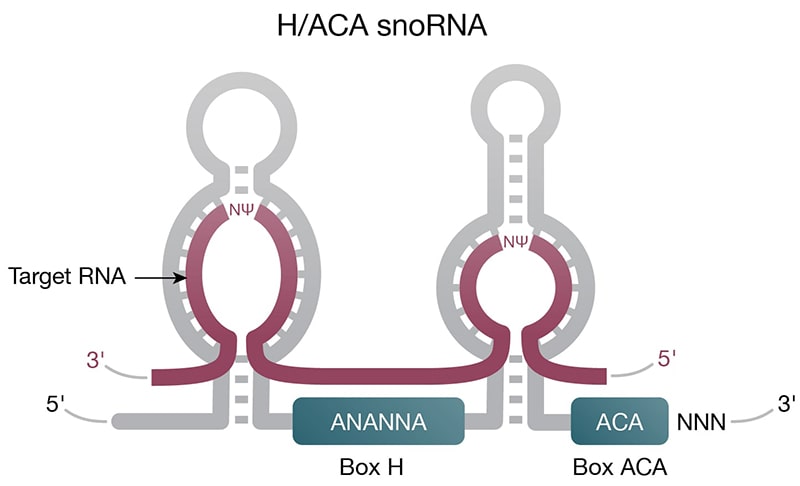
Figure 2. Structure of select ncRNAs. A) tRNA with three signature loops (grey, purple) and anticodon (shown as colored beads). B) Pre-miRNA with the region corresponding to mature miRNA (purple). C) Representation of long noncoding RNA (lncRNA) with complex secondary structure. D) H/ACA snoRNA (grey) has conserved H box (ANANNA) and ACA box (ACA) domains. Two hairpin loops contain the binding pocket for target RNA (purple) and are the sites of pseudouridylation (NΨ).
Long noncoding RNAs (lncRNA) (Fig 2D) are endogenous RNAs characterized by lengths longer than 200 nt, which allows them to fold into three-dimensional structures (Delpu 2016). As of March 2020, there are 18, lncRNA genes and 53,009 lncRNA transcripts deposited in the human GENCODE (Release 33) database. Similar to mRNA, lncRNAs are transcribed by RNA polymerase II and can be alternatively spliced, 5’ capped, and polyadenylated. There is also some overlap in their classification as noncoding RNA because one type of lncRNA, circular ncRNA (circRNA), is often derived from protein-coding mRNA transcripts. The lncRNAs are usually classified based on their genomic location, mechanism of action, or effect on DNA target. lncRNA expression levels are cell- and tissue–specific and shift with changes in cellular transcription, e.g., related to disease state or developmental stage. The largest class of lncRNAs are long intergenic noncoding RNAs (lincRNAs), which are not located near protein-coding genes (Ransohoff 2018).
Biological Roles of Noncoding RNAs
A surprising revelation of the Human Genome Project was that the roughly 20,000 protein-coding genes of the human body constitute only 1.5% of the human genome. While the noncoding bulk of the genome was presumed to lack any purpose, much of this “junk DNA” is actually transcribed into ncRNA and performs a range of vital biological functions (Chen 2015).
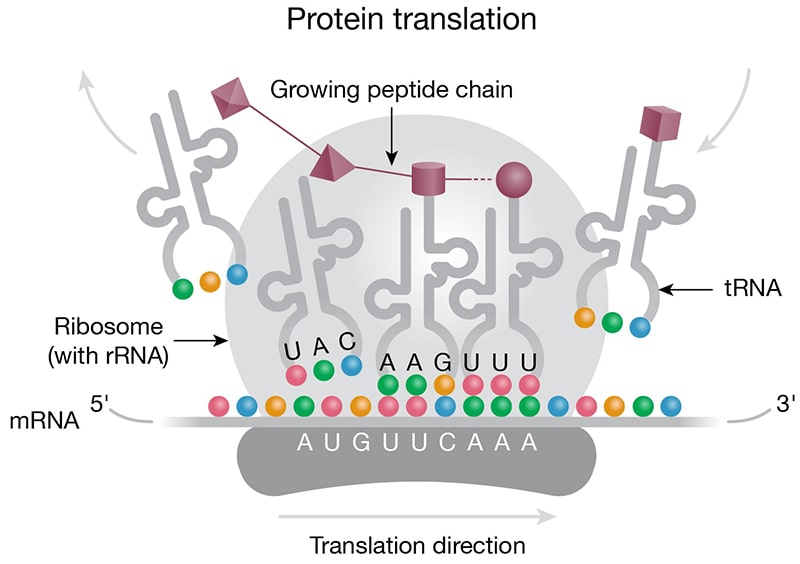
Housekeeping — Small nuclear RNAs (snRNAs) are abundant in the nucleus with important roles in intron splicing and RNA processing. In their natural form, snRNAs exist as ribonucleoprotein particles and, together with other proteins, form the spliceosome. Small nucleolar RNAs (snoRNAs) (Fig 2C) are found in the nucleolus or Cajal bodies, where they are part of ribonucleoprotein complexes involved in rRNA processing. While not small nucleotide length, rRNA and tRNA are also considered housekeeping nuclear ncRNAs with critical roles in mRNA translation (Fig 3A).
Figure 3
-
A
 B
B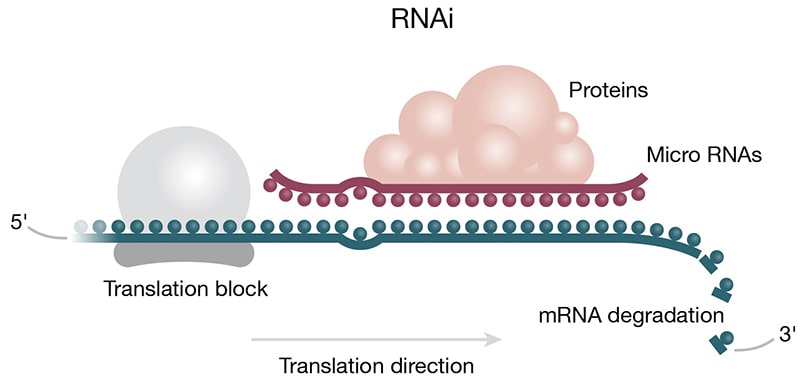
Figure 3. Roles of ncRNAs in protein translation. A) Protein translation machinery is assembled on the mRNA transcript. The ribosome moves along the mRNA transcript in the 5 to 3 prime direction. Anticodons (colored beads) on the tRNA base pair with the corresponding codons (colored beads) on the mRNA based on complementarity. The tRNAs deliver amino acids (purple 3-D shapes) to the elongating peptide chain (purple chain). B) Micro RNAs can act to regulate protein translation by degrading mRNA. Micro RNA complexed with proteins of the RISC complex (purple) is shown docked on the mRNA downstream of the translation block (blue/grey).
Gene Regulation — While gene regulation at the transcription step is well documented, ncRNAs can act to regulate gene expression by interacting with mRNA (Patil 2014). In the small ncRNA category, miRNA and siRNA act via the RNAi pathway and the RISC complex to inhibit translation (Fig 3B) (Phillips 2008). Circular ncRNAs (circRNAs) act as “miRNA sponges” to outcompete miRNAs in the cytoplasm; a popular example is CiRS-7, which inhibits oncogene regulator miR-7 (Kristensen 2019). Like miRNA, siRNA is generated by the action of the Dicer enzyme and joins the RISC complex to destabilize target mRNA molecules. However, siRNA is double-stranded and is considered exogenous RNA, meaning it is introduced by a foreign vector (e.g., a virus). Some plants and animals use siRNA silencing as an antiviral defense mechanism to weaponize viral RNA against itself, hence blocking viral propagation.
lncRNAs play a variety of roles in gene regulation at the level of both transcription and translation. For example, enhancer RNAs (eRNAs), which originate from enhancer regions, interact with neighboring promoters to enhance transcription by a mechanism that remains incompletely understood (Kim 2015). During 2010, a genome-wide sequencing study in mice, Kim et al. found that the eRNA expression levels at specific enhancers correlated with increased expression of close-proximity genes (Kim 2010). The largest group of lncRNAs, long intergenic ncRNAs (lincRNA), act in a cis or trans fashion to regulate gene expression (Ransohoff 2018). For example, HOTAIR is a trans-acting lincRNA that regulates chromatin structure and, when misregulated, acts epigenetically to upregulate oncogenes (Hajjari 2015).
Innate Immunity — lncRNAs are implicated in various facets of innate immunity, which is the initial line of defense against an infection (Hadjicharalambous 2019). This process begins with pathogen recognition, followed by an inflammatory response, and eventually pathogen removal. In a 2016 study, Kotzin et al. showed that the lncRNA Morrbid acts in a cis manner to regulate the survival of monocytes, neutrophils, and eosinophils via transcriptional regulation of a pro-apoptotic gene. Evidence suggests that IncRNAs are also involved in the maintenance of hematopoietic stems cells and activation of monocytes, macrophages, and dendritic cells (Hadjicharalambous 2019).
Adaptive Immunity — CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated protein) is an adaptive immunity mechanism used in bacteria to preserve DNA “memory” from invading viruses (Karvelis 2013). If the virus invades again, a targeting complex that includes guide RNA and Cas enzyme is used to target and cut the viral DNA. A key component of this neutralizing complex is a noncoding RNA called trans-activating CRISPR (tracr) RNA, which is used to form the guide RNA. The CRISPR-Cas system has been adapted as a tool for genome editing in research and clinical applications.
ncRNAs as Research Tools
RNAi — Although RNAi is not considered as a definitive antiviral mechanism in mammals, all the critical components of the pathway are evolutionarily conserved and present in mammalian cells. Thus, RNAi has been a valuable tool for gene knockdown studies in vitro and in vivo using exogenous siRNA (Mocellin 2004).
Antagomirs/Agomirs — Loss-of-function studies of miRNAs are an important way to better understand their diverse functions. Originally introduced in 2005, antagomirs are single-stranded, chemically engineered oligonucleotides that silence endogenous mRNAs (Krützfeldt 2005). In early studies, antagomir-122 was used to block murine miRNA-122, the most abundant miRNA in the liver, confirming its role in cholesterol biosynthesis (Esau 2006). Agomirs, conversely, are synthetic double-stranded miRNA mimetics that can be used in gene-function studies. Both molecules can be delivered in vitro via cell transfection, or in vivo via intravenous or local injection.
Diagnostic Applications of ncRNAs
Growing evidence suggests that the misregulation of ncRNAs is tied to human diseases, making them ideal candidates for diagnostic and therapeutic applications. In the small ncRNA category, miRNAs have shown utility in the clinic for diagnostic testing. In 2015, Montani et al. developed an mRNA signature blood test (miR-Test) for lung cancer screening (Montani 2015). miRNAs are also valuable markers for classifying cancer tumors and tissues of origin, which is critical for selecting the optimal treatment. A 2012 study used miRNA-based assays to identify 42 tumor types with high confidence (Meiri 2012). miRNA expression levels have also been associated with response to treatment in various cancers, suggesting the utility of miRNAs as indicators of resistance or otherwise suboptimal treatment outcomes (Wang 2019).
Studies over the past decade have demonstrated the utility of miRNAs and lncRNAs in diagnosing cancer; for example, using NGS sequencing to measure the expression of 10 miRNAs that are involved in thyroid cancer (Sistrunk 2019). This information is combined with sequencing data from a thyroid oncogene panel to detect cancer in thyroid nodules. lncRNAs are not as well characterized as miRNAs; however, reports of diagnostic applications are available. Overexpression of the prostate-specific lncRNA PCA3 is detected in prostate cancer cells; urine assays to detect PCA3 are commonly used in the clinic (Hessels 2009).
Therapeutic Applications of ncRNAs
Given their established roles in fundamental biology and human disease, ncRNAs are natural targets for therapeutic intervention. Many approaches have been used to restore or block miRNAs for a therapeutic purpose. Small molecules and oligonucleotide-based approaches (miRNA mimics) are documented ways to restore downregulated miRNAs (Ling 2013). The first restorative therapy candidate in clinical trial was a liposome-formulated miR34 mimic for advanced liver cancer — this trial was halted in 2016 due to reported adverse effects (Beg 2016). A miRNA mimic of the tumor suppressor miR-16, completed a Phase 1 clinical trial for the treatment of recurrent malignant pleural mesothelioma and non-small cell lung cancer (van Zandwijk 2017). Therapeutic strategies around blocking overactive miRNAs include artificial sponge RNAs, modeled after endogenous ncRNAs that act to soak up and block miRNAs.
siRNA therapies are centered around blocking mRNA using the RNAi pathway (Mahmoodi 2017). The first therapeutic effect using siRNA was reported in 2001, and the first clinical trial of an siRNA therapeutic was launched in 2004. Since then, 31 siRNA drug candidates have entered clinical trials for 18 diseases, including cancer, ocular, and metabolic disorders (Scherman 2017).
The first and second FDA-approved siRNA-based therapies target hereditary transthyretin-mediated (hATTR) amyloidosis (2018; ClinicalTrials.gov Identifier: NCT01829971) and the rare genetic disorder acute hepatic porphyria (2019; ClinicalTrials.gov Identifier: NCT02369198), respectively. A clinical trial initiated in 2008 used a nanoparticle delivery to stably deliver siRNA targeting the M2 subunit of ribonucleotide reductase (R2) to tumor cells via intravenous administration (Davis 2010). In a more recent clinical trial, siRNA packaged in a polymer capsule was used to target the KRAS oncogene (carrying G12D mutation) in solid pancreatic tumors (ClinicalTrials.gov Identifier: NCT01188785).
Ocular diseases have also been a favorite target of siRNA silencing therapies due to favorable attributes that allow siRNA drugs to be delivered easily and in high concentrations. For example, a 2012 clinical trial evaluated siRNA therapy for treating age-related macular degeneration (Nguyen 2012).
Related Topics and Products

Next-Generation Sequencing
Learn about next-generation sequencing (NGS) methods, the NGS workflow, and key technologies in the development of large-scale genomic sequencing.

Advances in RNAseq
NGS RNA sequencing (RNA-Seq) is the foremost tool for transcriptomics, gene expression profiling, and the study of differential RNA splicing.

RNA-Seq Workflow
A guide to the steps of an RNA-Seq workflow including library prep and quantitation and software tools for RNA-Seq data analysis.

PCR (Polymerase Chain Reaction)
Learn how PCR works, how to choose the right PCR instrument, how to design, optimize, analyze, and troubleshoot PCR assays.

Real-Time PCR (qPCR)
Discover an array of solutions to optimize, execute, and troubleshoot MIQE-compliant real-time PCR experiments.

Digital PCR (ddPCR)
Learn how breakthrough digital PCR technology is used in droplet digital PCR (ddPCR) to provide ultrasensitive and absolute nucleic acid quantitation.

lncRNA RT-qPCR Workflow
Bio-Rad's innovative workflow enables highly sensitive detection of long noncoding RNA (lncRNA) and helps to overcome challenges associated with IncRNA expression analysis.
Next-Generation Sequencing
Novel products for NGS library preparation and quantitation used in cutting-edge applications such as whole-transcriptome RNA-Seq and single-cell sequencing.

Traditional PCR Systems
DNA amplification instruments range from personal thermal cyclers to the flexible 1000-series.

qPCR Detection Systems
These systems deliver sensitive, reliable detection of both singleplex and multiplex real-time PCR reactions.

PCR & qPCR Reagents
A wide range of reagents for reverse transcription, PCR, and real-time PCR, optimized to generate accurate and reproducible data.

PCR Plastic Consumables
A large selection of thin-wall PCR tubes, plates, seals, and accessories precisely manufactured for optimal fit and cycling performance in Bio-Rad thermal cyclers, real-time PCR systems, ddPCR systems, and all major competitor systems.

PrimePCR PCR Primers, Assays & Arrays
Experimentally validated PCR primer and probe assays for gene expression, copy number variation, and mutation detection analysis for real-time PCR and Droplet Digital PCR.
Droplet Digital PCR Assays
Bio-Rad offers a comprehensive portfolio of Digital PCR Assays and Kits for numerous applications including Mutation Detection, Copy Number Determination, Genome Edit Detection, Gene Expression, Expert Design Assays, Residual DNA Quantification and Library Quantification.
References
- Beg MS et al. (2017). Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investigational new drugs, 35(2), 180–188. DOI: 10.1007/s10637-016-0407-y. PMCID: PMC5893501.
- Bhagavan NV, Ha C, Chapter 23 - RNA and Protein Synthesis, Editor(s): N.V. Bhagavan, Chung-Eun Ha, Essentials of Medical Biochemistry, Academic Press, 2011, Pages 301-320, ISBN 9780120954612, DOI: 10.1016/B978-0-12-095461-2.00023-0.
- Chen YA, Aravin AA. Noncoding RNAs in Transcriptional Regulation: The review for Current Molecular Biology Reports. Curr Mol Biol Rep. 2015 Mar 1;1(1):10-18. DOI: 10.1007/s40610-015-0002-6. PMCID: PMC4479201.
- Davis ME, Zuckerman JE, Choi CH, et at. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010 Apr 15;464(7291):1067-70. DOI: 10.1038/nature08956. PMCID: PMC2855406.
- Delpu Y, Larrieu D, Gayral M, et al. (2016) Noncoding RNAs: clinical and therapeutic applications (Chapter 12). In: Egger G (ed) Arimondo PBT-DD in CE. Academic Press, Boston, pp 305–326
- Esau C et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell. Metab. 3, 87–98 (2006). DOI: 10.1016/j.cmet.2006.01.005. PMID: 16459310.
- Hadjicharalambous MR, Lindsay MA. Long Noncoding RNAs and the Innate Immune Response. Noncoding RNA. 2019 Apr 19;5(2):34. DOI: 10.3390/ncrna5020034. PMCID: PMC6630897.
- Hajjari M, Salavaty A. HOTAIR: an oncogenic long noncoding RNA in different cancers. Cancer Biol Med. 2015 Mar;12(1):1-9. DOI: 10.7497/j.issn.2095-3941.2015.0006. PMCID: PMC4383848.
- Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009 May;6(5):255-61. DOI: 10.1038/nrurol.2009.40.
- Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013 May;10(5):841-51. DOIi: 10.4161/rna.24203. Epub 2013 Mar 27. PMCID: PMC3737341.
- Keam SP, Hutvagner G. tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression. Life (Basel). 2015 Nov 27;5(4):1638-51. DOI: 10.3390/life5041638. PMCID: PMC4695841
- Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010 May 13;465(7295):182-7. DOI: 10.1038/nature09033. PMCID: PMC3020079<./li>
- Kim TK, Hemberg M, Gray JM. Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect Biol. 2015 Jan 5;7(1):a018622. DOI: 10.1101/cshperspect.a018622.PMCID: PMC4292161.
- Kotzin JJ, Spencer SP, McCright SJ, et al. The long noncoding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016;537(7619):239–243. DOI: 10.1038/nature19346. PMCID: PMC5161578.
- Kowalski MP, Krude T. Functional roles of non-coding Y RNAs. Int J Biochem Cell Biol. 2015 Sep;66:20-9. DOI: 10.1016/j.biocel.2015.07.003. Epub 2015 Jul 6. PMCID: PMC4726728.
- Kristensen, L.S., Andersen, M.S., Stagsted, L.V.W. et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019. 20, 675–691. DOI: 10.1038/s41576-019-0158-7.
- Krützfeldt, J., Rajewsky, N., Braich, R. et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005. 438, 685–689. DOI: 10.1038/nature04303. PMID: 16258535.
- Ling H, Fabbri M, Calin GA. MicroRNAs and other noncoding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013 Nov;12(11):847-65. DOI: 10.1038/nrd4140. PMCID: PMC4548803
- Lodish H, Berk A, Zipursky SL, et al. Molecular Cell Biology. 4th edition. New York: W. H. Freeman; 2000. Section 4.4, The Three Roles of RNA in Protein Synthesis. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21603/
- Mahmoodi Chalbatani G, Dana H, Gharagouzloo E, Grijalvo S, Eritja R, et al. Small interfering RNAs (siRNAs) in cancer therapy: a nano-based approach. Int J Nanomedicine. 2019 May 2;14:3111-3128. DOI: 10.2147/IJN.S200253. PMCID: PMC6504672
- Meiri E, Mueller WC, Rosenwald S, et al. A second-generation microRNA-based assay for diagnosing tumor tissue origin. Oncologist. 2012;17(6):801-12. DOI:10.1634/theoncologist.2011-0466. PMCID: PMC3380879
- Mocellin S, Provenzano M. RNA interference: learning gene knock-down from cell physiology. J Transl Med. 2004 Nov 22;2(1):39. DOI: 10.1186/1479-5876-2-39. PMCID: PMC534783
- Montani F, Marzi MJ, Dezi F, et al. miR-test: a blood test for lung cancer early detection. J Natl Cancer Inst. 2015 107: 6. DOI: 10.1093/jnci/djv063
- Nguyen QD, Schachar RA, Nduaka CI, et al. MONET Clinical Study Group. Evaluation of the siRNA PF-04523655 versus ranibizumab for the treatment of neovascular age-related macular degeneration (MONET Study). Ophthalmology. 2012 Sep;119(9):1867-73. DOI: 10.1016/j.ophtha.2012.03.043.
- Alazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015 Jan 26;6:2. DOI: 10.3389/fgene.2015.00002. PMCID: PMC4306305.
- Patil VS, Zhou R, Rana TM. Gene regulation by noncoding RNAs. Crit Rev Biochem Mol Biol. 2014 Jan-49(1):16-32. DOI: 10.3109/10409238.2013.844092.PMCID: PMC4721600
- Phillips, T. (2008) Small noncoding RNA and gene expression. Nature Education 1(1):115 https://www.nature.com/scitable/topicpage/small-non-coding-rna-and-gene-expression-1078/
- Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic noncoding RNA. Nat Rev Mol Cell Biol. 2018 Mar;19(3):143-157. DOI: 10.1038/nrm.2017.104. PMCID: PMC5889127
- Scherman D, Rousseau A, Bigey P, et al. Genetic pharmacology: progresses in siRNA delivery and therapeutic applications. Gene Ther. 2017. 24, 151–156. DOI: 10.1038/gt.2017.6
- Sistrunk JW, Shifrin A, Frager M, et al. Clinical impact of testing for mutations and microRNAs in thyroid nodules. Diagnostic Cytopathology. 2019; 47: 758– 764. DOI: 10.1002/dc.24190
- Van Zandwijk N, Pavlakis N, Kao SC, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol 2017;18:1386-96. DOI: 10.1016/S1470-2045(17)30621-6
- Wang W, Han C, Sun Y, et al. Noncoding RNAs in cancer therapy resistance and targeted drug development. J Hematol Oncol 12, 55 (2019). DOI: 10.1186/s13045-019-0748-z
