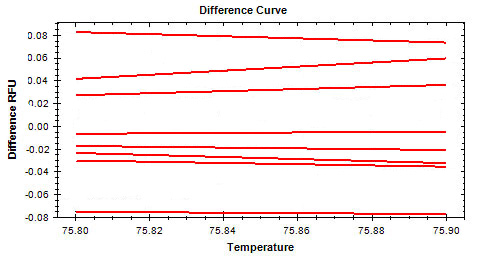
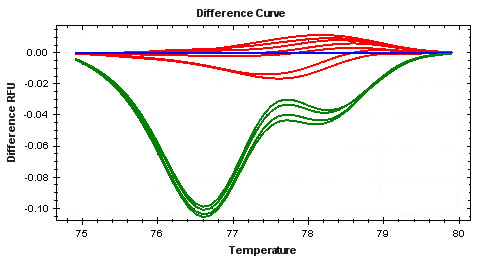
Listed below are some common symptoms in an HRM experiment. Select one of the symptoms to view possible causes and solutions.
Related Topics: Gene Expression/Quantification Experiments and Allelic Discrimination Experiments.
Symptoms
Missed Call or Clustering
Back to TopA missed call or clustering may be due to the causes below. Select one of the causes for possible solutions.
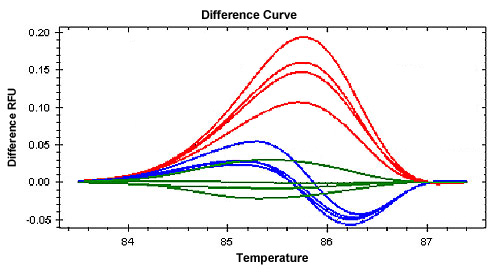
Small Thermal Changes
Back to Top|
Small thermal changes between two sequences can be difficult to differentiate, and these changes can arise from sources such as sequence differences, sample quality, amplicon length, primer design, and PCR reagents. |
|
| Possible Solutions | Use at least one positive control for each target. |
| Decrease the amplicon size. This will increase the difference in signal at a given temperature between two sequences that differed at only one nucleotide position. | |
| During analysis, use the cluster with the most distinct visual difference between the various sequence types as the reference cluster. This will help minimize trace overlap. | |
| Perform a dilution series of the PCR template to determine whether the effect of the inhibitory agent can be reduced. | |
Recommended Products and Links
- Precision Melt Supermix
- Precision Melt Analysis Software
- DNA Isolation
- RNA Isolation
- Aurum™ Total RNA Kits
- PureZOL™ RNA Isolation Reagent
Primer Design
Back to Top|
In some cases, the GC content of the primers can affect the melting temperature of your target. Ensuring that the temperature shift will not be affected by the sequence of your primer set will improve calls and clustering. |
|
| Possible Solutions | Design new primers with less GC content. |
Amplification Did Not Reach Plateau
Back to TopLow-Quality DNA
Back to Top|
Low-quality DNA may prevent samples from reaching high plateau, which can result in inconclusive high resolution melt (HRM) data. Improving the DNA quality will ensure that the HRM data is not skewed and will be called appropriately. |
|
| Possible Solutions | Increase starting quantity. |
| Add more PCR cycles to protocol. | |
Long Amplicon
Back to Top|
The length of an amplicon can influence the amplification and melt profile if the sequence contains more than one mutation. As a result, the mutation of interest can be difficult to separate from the other sequence variants if primer sets are designed for longer amplicons (> 300 bp). |
|
| Possible Solutions | Use amplicons with a target length <300 bp to improve detection of sequence variants. |
Recommended Products and Links
Poor Amplification or Failed Reaction
Back to TopLow-Quality DNA
Back to Top|
Successful HRM analysis is highly dependent on the quality of the amplicons being compared. Low-quality DNA may produce nonspecific PCR products that can affect the melt profile. |
|
| Possible Solutions | Increase starting concentration. |
| Add more PCR cycles to protocol. | |
Inhibitors
Back to Top|
Sometimes inhibitors of PCR are carried over from sample preparation (nucleic acid extraction). Common PCR inhibitors include phenol, detergents, proteases, and residual compounds from source materials, such as animal or plant tissue, body fluids, or soil preparations. Suboptimal reactions containing inhibitors are likely to affect the reactions and thus any postanalysis, such as HRM analysis. |
|
| Possible Solutions | Take extra care with the nucleic acid extraction steps to minimize carryover of PCR inhibitors. |
| Add more PCR cycles to protocol. | |
Recommended Products and Links
- Precision Melt Supermix
- DNA Isolation
- RNA Isolation
- Aurum™ Total RNA Kits
- PureZOL™ RNA Isolation Reagent
Further Reading

Castellanos E et al. (2010). Rapid identification and differentiation of Mycobacterium avium subspecies paratuberculosis types by use of real-time PCR and high-resolution melt analysis of the MAP1506 locus. J Clin Microbiol 48, 1474–1477. PMID: 20129970
Gan XL et al. (2010). Association of an interleukin-16 gene polymorphism with the risk and pain phenotype of endometriosis. DNA Cell Biol 29, 663–667. PMID: 20662556
Martino et al. (2010). Application of high-resolution melting to large-scale, high-throughput SNP genotyping: A comparison with the TaqMan method. J Biomol Screen 15, 623–629. PMID: 20371868
Temesvári M et al. (2011). High-resolution melting curve analysis to establish CYP2C19*2 single nucleotide polymorphism: Comparison with hydrolysis SNP analysis. Mol Cell Probes 25, 130–133. PMID: 21315147
Wu W et al. (2011). Examination of AKAP12 promoter methylation in skin cancer using methylation-sensitive high-resolution melting analysis. Clin Exp Dermatol 36, 381–385. PMID: 21198787
Yu B et al. (2011). Polymorphisms of PXK are associated with autoantibody production, but not disease risk, of systemic lupus erythematosus in Chinese mainland population. Lupus 20, 23–27. PMID: 20829310