Droplet Digital PCR (ddPCR) is a method for performing digital PCR that is based on water-oil emulsion droplet technology. A sample is fractionated into 20,000 droplets, and PCR amplification of the template molecules occurs in each individual droplet. ddPCR technology uses reagents and workflows similar to those used for most standard TaqMan probe-based assays. The massive sample partitioning is a key aspect of the ddPCR technique.
This section provides an overview of droplet digital PCR technology, how ddPCR works, and what the advantages and benefits of ddPCR are.
Related topics: Planning Droplet Digital PCR Experiments, Absolute Quantification of PCR Targets with the Droplet Digital™ PCR System

Validated ddPCR Assays
Our PrimePCR™ Assays for Droplet Digital™ PCR are designed and validated for copy number variation, rare mutation detection, NGS library quantification, and high-sensitivity gene expression studies.
Learn more »Page Contents
Droplet Digital PCR technology is a digital PCR method utilizing a water-oil emulsion droplet system. Droplets are formed in a water-oil emulsion to form the partitions that separate the template DNA molecules. The droplets serve essentially the same function as individual test tubes or wells in a plate in which the PCR reaction takes place, albeit in a much smaller format. The massive sample partitioning is a key aspect of the ddPCR technique.
The Droplet Digital PCR System partitions nucleic acid samples into thousands of nanoliter-sized droplets, and PCR amplification is carried out within each droplet. This technique has a smaller sample requirement than other commercially available digital PCR systems, reducing cost and preserving precious samples.
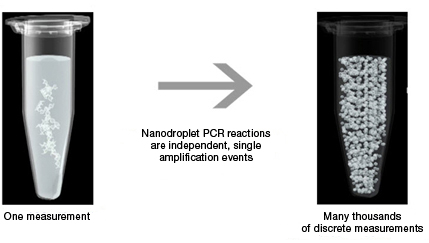
Sample partitioning is the key to droplet digital PCR. In traditional PCR, a single sample offers only a single measurement, but in Droplet Digital PCR, the sample is partitioned into 20,000 nanoliter-sized droplets. This partitioning enables the measurement of thousands of independent amplification events within a single sample.
ddPCR technology uses a combination of microfluidics and proprietary surfactant chemistries to divide PCR samples into water-in-oil droplets (Hindson et al. 2011). The droplets support PCR amplification of the template molecules they contain and use reagents and workflows similar to those used for most standard TaqMan probe-based assays. Following PCR, each droplet is analyzed or read to determine the fraction of PCR-positive droplets in the original sample. These data are then analyzed using Poisson statistics to determine the target DNA template concentration in the original sample.
Droplet Digital PCR surpasses the performance of earlier digital PCR techniques by resolving the previous lack of scalable and practical technologies for digital PCR implementation. Serial dilution is laborious and introduces the possiblity of pipetting error; competing chip-based systems rely on complex fluidics schemes for partitioning. Droplet Digital PCR addresses these shortcomings by massively partitioning the sample in the fluid phase in one step. The creation of tens of thousands of droplets means that a single sample can generate tens of thousands of data points rather than a single result, bringing the power of statistical analysis inherent in digital PCR into practical application. Bio-Rad's Droplet Digital PCR System automates the ddPCR workflow of droplet generation, thermal cycling, droplet reading, and data analysis, making this technology accessible to the working research laboratory.
ddPCR technology enables high-throughput digital PCR in a manner that uses lower sample and reagent volumes and reduces overall cost compared with other methods while maintaining the sensitivity and precision that are the hallmarks of digital PCR.
The benefits of ddPCR technology include:
- Absolute quantification — ddPCR technology provides an absolute count of target DNA copies per input sample without the need for running standard curves, making this technique ideal for measurements of target DNA, viral load analysis, and microbial quantification
- Unparalleled precision — the massive sample partitioning afforded by ddPCR enables the reliable measurement of small fold differences in target DNA sequence copy numbers among samples
- Increased signal-to-noise ratio — high-copy templates and background are diluted, effectively enriching template concentration in target-positive partitions, allowing for the sensitive detection of rare targets and enabling a ±10% precision in quantification
- Removal of PCR bias — error rates are reduced by removing the amplification efficiency reliance of qPCR, enabling the detection of small (1.2-fold) differences
- Simplified quantification — neither calibration standards nor a reference (the ΔΔCq method) is required for absolute quantification
- Reduced consumable costs — reaction volumes are in the pico- to nanoliter ranges, reducing reagent use and the sample quantity required for each data point
- Lower equipment costs — the emulsion-based reaction system means that the PCR reactions can be performed in a standard thermal cycler without complex chips or microfluidics
- Superior partitioning — ddPCR technology yields 20,000 droplets per 20 µl sample, nearly two million partitioned PCR reactions in a 96-well plate, whereas chip-based digital PCR systems produce only hundreds or thousands of partitions. The greater number of partitions yields higher accuracy
Bio-Rad’s QX200 Droplet Digital PCR (ddPCR) System consists of two instruments, the QX200 Droplet Generator and the QX200 Droplet Reader, plus their associated software and consumables. The QX200 Droplet Generator partitions samples (20 µl into 20,000 nanoliter-sized droplets) for PCR amplification. Following amplification using a thermal cycler, droplets from each sample are analyzed individually on the QX200 Droplet Reader, where PCR-positive and PCR-negative droplets are counted to provide absolute quantification of target DNA in digital form.
The ddPCR System can be used to:
- Detect rare DNA target copies with unmatched sensitivity
- Determine copy number variation with unrivaled accuracy
- Measure gene expression levels with exquisite precision
QX200 Droplet Digital PCR System. QX200 Droplet Reader (left) and the QX200 Droplet Generator (right).
This 3-D animated video describes the principles and process of ddPCR using Bio-Rad's Droplet Digital PCR System.
The workflow is quite simple. First, the QX200 Droplet Generator partitions samples into thousands of nanoliter-sized droplets. After PCR on a thermal cycler, droplets from each sample are streamed in single file on the QX200 Droplet Reader to count positive and negative reactions. The PCR-positive and PCR-negative droplets are counted to provide absolute quantification in digital form. The steps are described in more detail below.
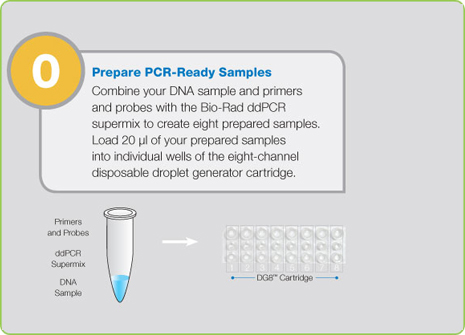
Step 0: Prepare PCR-Ready Samples prior to Starting ddPCR

Step 1: Droplet Generation
Prior to droplet generation, nucleic acid samples (DNA or RNA) are prepared as they are for any real-time assay: using primers, fluorescent probes (TaqMan probes with FAM and HEX or VIC), and a proprietary supermix developed specifically for droplet generation. Samples are then placed into the QX200 Droplet Generator, which utilizes proprietary reagents and microfluidics to partition the samples into 20,000 nanoliter-sized droplets. The droplets created by the QX200 Droplet Generator are uniform in size and volume.
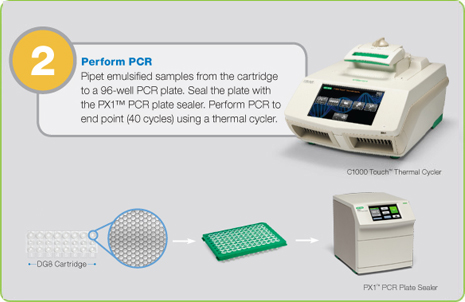
Step 2: PCR Amplification of Droplets
Droplets are transferred to a 96-well plate for PCR amplification in any compatible thermal cycler.
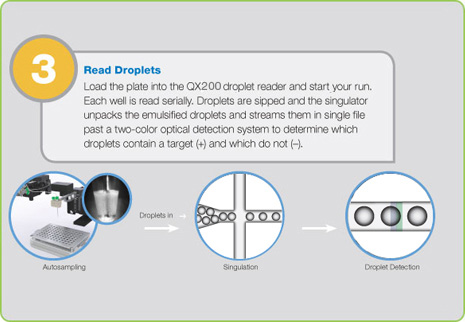
Step 3: Droplet Reading
Following PCR amplification of the nucleic acid target in the droplets, the samples are placed in the QX200 Droplet Reader, which analyzes each droplet individually using a two-color detection system (set to detect FAM and either HEX or VIC), enabling multiplexed analysis for different targets in the same sample. The droplet reader and its bundled QuantaSoft™ software count the PCR-positive and PCR-negative droplets.
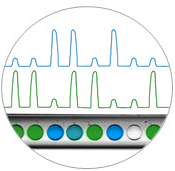
Digital droplet reading. Fluorescence measurements for each droplet in two optical channels are used to count the numbers of positive and negative droplets per sample.

Step 4: Analyze Results
Positive droplets, containing at least one copy of the target, exhibit increased fluorescence over negative droplets. In ddPCR, the QuantaSoft software measures the numbers of droplets that are positive and negative for each fluorophore (for example, FAM and HEX) in a sample. The fraction of positive droplets is then fitted to a Poisson distribution to determine the absolute initial copy number of the target DNA molecule in the input reaction mixture in units of copies/µl.
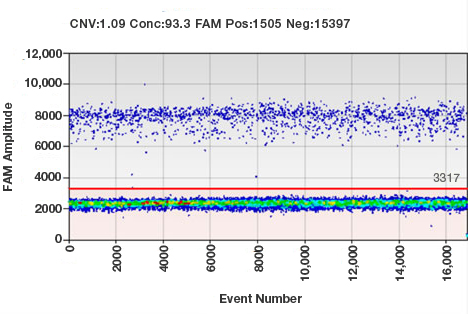
Sample results from a ddPCR experiment. Each droplet in a sample is plotted on a graph of fluorescence intensity versus droplet number. All positive droplets (those above the threshold intensity indicated by the red line) are scored as positive, and each is assigned a value of 1. All negative droplets (those below the threshold) are scored as negative, and each is assigned a value of 0 (zero). This counting technique provides a digital signal from which to calculate the starting target DNA concentration by a statistical analysis of the numbers of positive and negative droplets in a given sample.
Droplet Digital PCR data from a multiplex experiment in which two targets are PCR amplified can also be viewed as a 2-D plot in which FAM fluorescence is plotted versus HEX fluorescence for each droplet. Because the DNA distribution into the droplets follows a random pattern, the droplets are clustered into four groups:
- FAM-negative, HEX-negative
- FAM-positive, HEX-negative
- FAM-negative, HEX-positive
- FAM-positive, HEX-positive
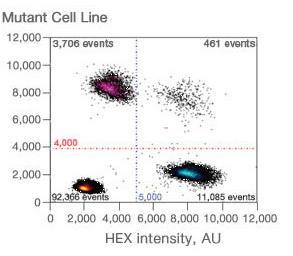
2-D plot of droplet fluorescence.
The QuantaSoft software fits the fraction of positive droplets to a Poisson distribution to determine the absolute starting copy number in units of copies/µl input sample and then reports the target DNA concentration in the form of copies per µl in the sample.
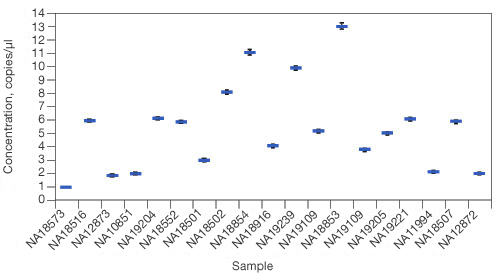
Concentration results. Sample concentrations are plotted as copies per µl.
Step 5: Visualize Data
- Bizouarn F http://www.genengnews.com/gen-articles/digital-pcr-improving-nucleic-acid-quantification/4093/
- Hindson BJ et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83(22): 8604–8610.
- Pinheiro LB et al. (2012). Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 84, 1003–1011
- Third Generation PCR (http://www.bioradiations.com/focus-on-technology/62-pcr/1329-third-generation-pcr)
- Droplet Digital PCR Opens New Perspectives in HIV Research (http://www.bioradiations.com/focus-on-technology/783-ddpcr/1356-droplet-digital-pcr-opens-new-perspectives-in-hiv-research-)
- A Simple Method for Obtaining Absolute Quantification of DNA Molecules using the Innovative Droplet Digital PCR Technology (http://www.bioradiations.com/focus-on-technology/783-ddpcr/1340-simpleddpcr)
- A New Paradigm for Precise Quantitation of RNA (http://www.bioradiations.com/focus-on-technology/783-ddpcr/1333-ddpcr)
- Sykes PJ et al. (1992). Quantitation of targets for PCR by use of limiting dilution. Biotechniques 13: 444–449. PMID: 1389177
