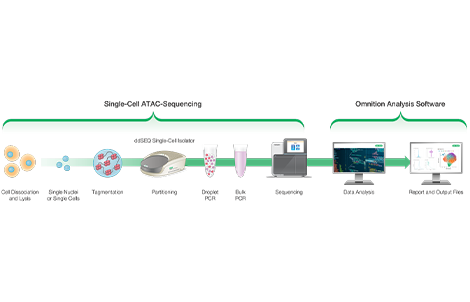
Description
Description
Download Omnition Analysis Software
The Omnition Analysis Software Version 1.0.0 (ATAC module) is a pipeline analysis tool that can QC, analyze and report single-cell ATAC-Seq data generated from the Bio-Rad SureCell ATAC-Seq assay and ddSEQ-deployed combinatorial indexing ATAC-Seq protocols as described in Lareau et al. (2019).
Compared to the Bio-Rad ATAC Analysis Toolkit, Omnition Analysis Software offers better usability and functionality as highlighted below:
- Streamlined, robust and efficient workflow
- Enabling batch analysis and reporting for multiple samples
- Supporting ddSEQ-deployed combinatorial indexing ATAC protocols
- Open-source, available for download
Applications and Uses
- Single-cell ATAC-Seq data analysis and reporting
- Single-cell combinatorial indexing ATAC-Seq data analysis and reporting
How to Run the Omnition
Omnition v1.0.0, the ATAC module, enables QC, analysis and reporting of data generated with the ddSEQ-deployed single-cell ATAC assay and combinatorial indexing ATAC protocols. Watch this tutorial video to learn how to install the Omnition and how to process and report your single-cell ATAC-seq data.
Example Omnition Reports
- Single-cell ATAC-Seq data analysis report
- Single-cell combinatorial indexing ATAC-Seq data analysis report
Data Sets*
- Dataset 1 — Mouse Brain (46,653 cells): Frozen whole brain tissue from two adult mice, purchased from BrainBits, and processed according to the Omni-ATAC Protocol to isolate nuclei.
- Dataset 2 — Combinatorial profiling of resting and stimulated bone marrow (136,463 cells): Frozen human bone marrow mononuclear cells from AllCells, stimulated with and without LPS, and barcoded with 96 indexed Tn5 transposases prior to droplet encapsulation.
- Dataset 3 — Isolated populations from peripheral blood (52,873 cells): Frozen human bone marrow CD34+ stem/progenitor cells, and isolated peripheral blood CD4+ T cells, CD8+ T cells, Monocytes, B cells, and NK cells, bought from AllCells and processed according to the Whole-Cell Protocol provided in the ATAC-Seq User Guide.
*Datasets are from SubSeries that are part of the SuperSeries cited below.
SuperSeries: Lareau CA, Duarte FM, Chew JG, Kartha VK et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat Biotechnol 2019 Aug;37(8):916-924.
