The ProteOn™ XPR36 protein interaction array system uses surface plasmon resonance (SPR) technology to monitor biomolecular interactions in real time without the use of labels. Changes in mass at the sensor chip surface are detected as a change in the SPR signal; hence, an SPR biosensor can detect the binding of an analyte molecule flowing in solution over an immobilized target. SPR technology is used for a wide range of applications in basic biological research and can detect binding interactions between many different types of molecules such as antibodies/antigens, small molecules/protein targets, and whole cells/lipid membranes. Because SPR is a time-resolved method, as opposed to endpoint assays such as ELISA or isothermal calorimetry (ITC), data are collected in real time during both the association and dissociation phases, allowing for quantification of kinetic rate constants. Knowing the kinetic parameters of an interaction allows for easy discrimination between tight and weak binders and leads to improved design in drug discovery. Thermodynamic data about the energetics of binding can also be obtained from SPR by studying the binding kinetics of an interaction at multiple temperatures.
Related Topics: Large and Small Molecule Screening by SPR and SPR Assay Development and Experimental Optimization.
Page Contents
The unique 6 x 6 fluidics of the ProteOn XPR36 system deliver robust kinetics for up to six targets in one shot. Up to 36 different interactions can be monitored simultaneously by injecting six different analytes over six different ligands. This high-throughput feature of the ProteOn system allows for fast and accurate characterization of multiple binding interactions in parallel, reducing sample processing time and greatly speeding protocol optimization. The novel XPR™ technology confers advantages found only on the ProteOn system, such as real-time double referencing — which allows for highly stable baselines — and increased flexibility in experimental design (Bronner et al. 2006, Cohen et al. 2009, Cohavi et al. 2009). Association (ka) and dissociation (kd) kinetic constants can be easily extracted from the data by using the ProteOn Manager™ Software to fit experimental data to a number of binding models, ranging from a simple Langmuir 1:1 binding model to Langmuir binding with mass transport or heterogeneous ligand and analyte models.
Webinars addressing various aspects of Kinetic Characterization with the ProteOn XPR36 system:
ProteOn XPR36 and Lipoparticle Technology, a Powerful Combination for Screening Antibody Therapeutics Against Membrane Proteins
Presented by Sharon Willis, PhD (Integral Molecular, USA)
Download recorded webinar
Novel Liposome Immobilization Using LayerLab Technology
Presented by Torbjorn Pettersson, PhD and Magnus Branden, PhD (LayerLab, Sweden)
Download recorded webinar
Pushing ProteOn to Its Limits — From Protein-Protein Docking to Nanoparticle Interactions
Presented by Gideon Schreiber, PhD (Weizmann Institute, Israel)
Download recorded webinar
Bio-Rad is Evolving SPR — Featuring the New ProteOn Manager Software and the New His-tag Capture Sensor Chip
Presented by Ruben Luo, PhD (Bio-Rad Laboratories, USA)
Download recorded webinar
New Features of ProteOn Manager v3.0 and HTG Sensor Chip (English & Mandarin)
Presented by: Ruben Luo, PhD (Bio-Rad Laboratories)
Download recorded webinar
Knowing the thermodynamics of biomolecular interactions, in addition to the kinetics, can lead to a better understanding of the mechanisms behind binding and help improve rational drug design. SPR technology is capable of measuring the energetics of binding by both equilibrium-based and kinetics-based analyses. Typical thermodynamic binding experiments involve measuring kinetic parameters with the ProteOn system under a range of temperatures and calculating the dissociation equilibrium constant (KD) at each temperature. The enthalpy and entropy of binding can then be extracted by fitting the experimental data to the van't Hoff equation. Kinetic parameters measured at different temperatures with the ProteOn system can also be used to obtain transition state thermodynamics, which can be useful in designing higher affinity ligands. The ability of the ProteOn system to provide high-quality and high-throughput kinetic data at different temperatures enables parallel thermodynamic analysis of multiple biomolecular interactions (Bravman et al. 2006 and 2008).

Model system investigated by John Kulman (see webinar for details).
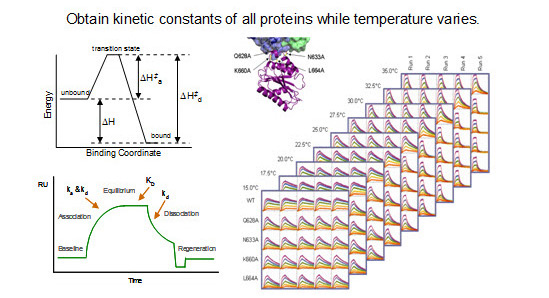
Thermodynamic data is determined.
Webinar relevant to thermodynamics of biomolecular interactions:
A Calcium-Dependent Immunocapture Strategy for Enhanced-Throughput SPR
Presented by John Kulman, PhD (Puget Sound Blood Center, USA)
Click to download recorded webinar: https://biorad.box.net/shared/j00jbuzdet
SPR technology serves as an efficient and powerful platform for the quantitation of biological samples. Protein quantitation and concentration analysis can be used for quality control of downstream processing or for detecting biomarkers in diagnostic research. Two major advantages of SPR over other labeled techniques (such as ELISA) for concentration analysis are that (1) label-free SPR eliminates the effort involved in labeling the analyte of interest and (2) using an SPR sensor chip surface with binding specificity (either by using a capture agent or immobilizing ligands to the surface) allows for the direct analysis of molecules from crude samples without prior sample purification.
Protein quantitation is achieved either by measuring the response at equilibrium (similar to ELISA but label free) or by obtaining the initial binding rate. The latter is often used due to its ease of use and rapid results. The concentration of an unknown sample is calculated by comparing the binding response under these conditions to a standard curve of binding responses for known concentrations. The parallel fluidics and reproducibility of the ProteOn system enables reliable and high-throughput concentration analysis of biological samples for both research applications and manufacturing and quality control processes.
The figures and table below illustrate the protein quantitation capabilities of the ProteOn Manager software. Data can be managed in a tabular or graphical format to display the concentration values from the standards, unknowns and control samples.
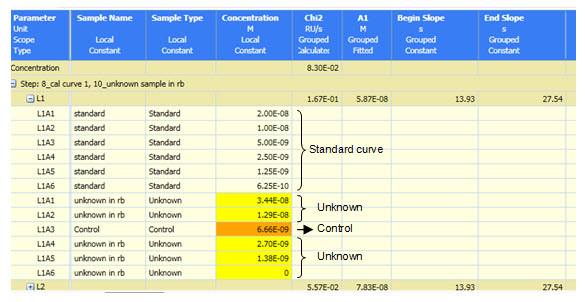
Example of protein quantitation data.
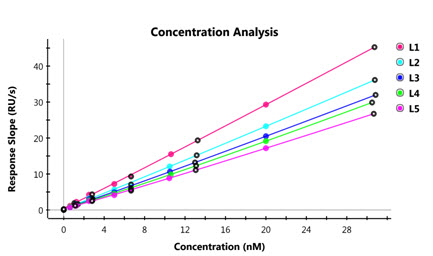
Example of standard curve.

Example of protein quantitation analysis.
Bravman T et al. (2006). Exploring "one-shot" kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal Biochem 358, 281–288. PMID: 16962556
This article describes the use of the ProteOn™ XPR36 parallel array biosensor to characterize the binding kinetics of a set of small molecule–enzyme interactions. Using the ProteOn system’s crisscrossing flowpath system, the authors collected response data for six different concentrations of each analyte over six different target protein surfaces. This "one-shot" approach to kinetic analysis significantly improves throughput while generating high-quality data even for low molecular mass analytes. The temperature dependence of the kinetics for four of the inhibitor-enzyme interactions was also determined in the range of 15–35°C. These results illustrate the potential of this new parallel-processing biosensor to increase the speed of kinetic analysis in drug discovery and expand the applications of real-time protein interaction arrays.
Bravman T et al. (2008). The ProteOn XPR36 array system — high-throughput kinetic binding analysis of biomolecular interactions. Cell Mol Bioeng 1, 216–228.
This article describes how the ProteOn system was used to investigate the interactions between carbonic anhydrase isozyme (CAII) and a series of known small molecule sulfonamide inhibitors. Kinetic screening results were obtained quickly using the One-shot Kinetics™ approach. The measured affinity of nine inhibitors were consistent with those determined using isothermal titration calorimetry. The temperature dependence of the kinetics was measured, and van't Hoff plots were obtained to determine the thermodynamics of the CAII- carboxybenzenesulfonamide (CBS) interaction.
Bronner V et al. (2006). Mechanisms of protein-protein binding: double-mutant cycle analysis using the ProteOn™ XPR36 system. Bio-Rad Bulletin 5358.
This study employed the ProteOn system to uncover important residues for the interaction of two proteins, TEM1 beta-lactamase and its inhibitor beta-lactamase inhibitor protein (BLIP). A double-mutant cycle (DMC) analysis was used along with the innovative One-shot Kinetics™ of the ProteOn system. The DMC analysis is an excellent tool to investigate the structure, mechanism, and dynamics of protein-protein interactions. Multiple mutants of both TEM1 and BLIP were analyzed against each other to determine the contribution of residues toward the stability of the TEM1-BLIP interaction.
Cohavi O et al. (2009). Docking of antizyme to ornithine decarboxylase and antizyme inhibitor using experimental mutant and double-mutant cycle data. J Mol Biol 390, 503–515. PMID: 19465028
This article employs the ProteOn™ system to analyze the interaction between ornithine decarboxylase (ODC) and a regulatory protein antizyme (Az). The binding sites of ODC on Az were mapped using high-throughput mutagenesis and computational docking. Double-mutant cycle (DMC) analysis between residues on Az and ODC was used to obtain further insights on the structure and function of the complexes.
Cohen S et al. (2009). Applications of the ProteOn™ NLC sensor chip: antibody-antigen, DNA-protein, and protein-protein interactions. Bio-Rad Bulletin 5449.
This tech note describes the specificity and stability of biotinylated ligands captured to the ProteOn and NLC sensor chip. Three common biomolecular interaction models representing antigen-antibody, DNA-protein, and protein-protein interactions were analyzed using the One-shot Kinetics™ approach. These three interaction types were analyzed in high throughout with the 6 x 6 interaction array. It also allowed for flexibility in experimental design.
Yousef M (2007). Advances in rapid monoclonal antibody screening. Am Biotechnol Lab 25, 26–28.
This article describes an alternative method for the rapid screening of monoclonal antibodies using multiplexed SPR and the One-shot Kinetics™ approach of the ProteOn™ XPR36 protein interaction array system. The ProteOn system was used to screen supernatants to identify high-affinity mAb candidates against human IL-12 and hemoglobin E. Over 250 supernatants were screened in 12.5 hr in one experiment, using a single sensor chip. There was no need to purify antibodies from the supernatants prior to analysis.
Related Content
TEST
General ProteOn Literature
| 5390 | ProteOn — PIA XPR36 Brochure | Click to download |
| 5538 | ProteOn XPR36 — Analyzing Protein Interaction Array System Featured Article Reprint | Click to download |
| 5413 | ProteOn XPR36 Hardware — 36 Interactions on a Single Chip: Label-Free, in Real-Time Product Information Sheet | Click to download |
| 5627 | ProteOn Manager Software Product Information Sheet | Click to download |
| 5404 | ProteOn Sensor Chips — Application-Specific Surface Chemistries = Optimized Ligand Activty Product Information Sheet | Click to download |
| 5410 | ProteOn Protocol Development Kits Product Information Sheet | Click to download |
| 5409 | Protein Interaction Analysis — ProteOn XPR36 System Ordering Information Sheet | Click to download |
| 6414 | ProteOn™ XPR36 Experimental Design and Application Guide, Rev B | Click to download |
Large Molecule Protein-Interaction Analysis
| 3172 | ProteOn PIA — Rapid and Efficient Determination of Kinetic Rate Constants using the ProteOn XPR36 Protein Interaction Array System | Click to download |
| 5412 | ProteOn App Guide — Antibody Characterization and Development Using the ProteOn XPR36 PIA System Application Guide Product Information Sheet | Click to download |
| 5540 | ProteOn — PIA Screening, Ranking, and Epitope Mapping of Anti-Human IL-9 Supernatants | Click to download |
| 5368 | ProteOn PIA — Analysis of Multiple Protein-Protein Interactions Using the ProteOn XPR Protein Interaction Array System | Click to download |
| 5449 | Protein Interaction Analysis Applications of the ProteOn NLC Sensor Chip: Antibody-Antigen, DNA-Protein-Protein Interactions | Click to download |
| 5358 | ProteOn PIA — Mechanisms of Protein Binding: Double-Mutant Cycle Analysis Using the ProteOn XPR36 System | Click to download |
| 5820 | ProteOn — Rapid Screening and Selection of Optimal Antibody Capturing Agents Using the ProteOn XPR36 Protein Interaction Array System | Click to download |
| 5360 | ProteOn PIA — Rapid and Detailed Analysis of Muliple Antigen-Antibody Pairs Using the ProteOn XPR36 Protein Interaction Array System | Click to download |
| 5367 | ProteOn PIA — Rapid Optimization of Immobilization and Binding Conditions for Kinetic Analysis of Protein-Protein Interactions using the ProteOn XPR36 PI Array System | Click to download |
Small Molecule Analysis
| 5965 | Rapid High-Throughput Screening of Protein Kinase Inhibitors Using the ProteOn PIA System | Click to download |
| 5797 | Protein Interaction — Rapid Assay Development and Optimization for Small Molecule Drug Discovery | Click to download |
| 5679 | ProteOn — Applications of the ProteOn GLH Sensor Chip: Interaction Between Proteins and Small Molecules | Click to download |
| 5960 | High-Throughput Profiling of Kinase Inhibitors Selectivity Using the ProteOn XPR36 Protein Interaction Array System | Click to download |
| 5846 | Determining the Binding Kinetics of HIV-1 Nucleocapsid Protein to Six Densities of Oligonucleotide Using the ProteOn XPR36 Protein Interaction Array System | Click to download |
| 5822 | How to Perform Excluded Volume Correction on the ProteOn XPR36 Protein Interaction System Product Guide | Click to download |
