The characterization of stem cell cultures has two main purposes: monitoring the genomic integrity of the cells and tracking the expression of proteins associated with pluripotency.
Genomic analysis is necessary to ensure stem cells maintained in culture have not undergone chromosomal changes through chromosomal loss or duplication, or changes in their epigenetic profiles. Proteomic analysis ensures that the cells are expressing the factors necessary to maintain pluripotency.
Confirmation of the differentiated state by analyzing key genetic and protein markers ensures identification and propagation of the correct cell type. This section provides an overview of different analysis methods for stem cells including karyotyping, single-nucleotide polymorphism (SNP) analysis, epigenetic profiling, flow cytometry and immunocytochemistry, RT-qPCR, digital PCR, western blotting, biomarker analysis, and teratoma formation.
Related Topics: Isolation and Maintenance of Stem Cells, Differentiation of Stem Cells, Transfection and Transduction of Stem Cells, and Stem Cells in Therapeutics and Research.
Page Contents
A variety of factors affect stem cell cultures
Factors
Media composition
Cell density
Feeder cell type/density
Growth factors/additives
Feeder free culture
Passage method
Number of passages
Freezing and thawing protocols
Microbial contamination
Possible changes
Chromosomal
Phenotype/morphology
Differentiation
Pluripotency loss
Epigenetic changes
Tumorigenesis
Loss of self-renewal ability
Stem cell cultures must be periodically assessed for changes in morphology, karyotype, and ability to differentiate.
Adaptation to culture conditions and maintenance in culture of any cell type will change the population. As cells divide and are passaged, the composition of the population changes due to selective pressures on the cells. Additionally, cells that grow and divide more rapidly will become dominant, gradually changing the composition of stem cell populations in culture. Mosaic populations often develop, with several distinct populations evolving. Therefore, keeping stocks of early passages of cells and continual monitoring are important.
A number of techniques are used for genetic characterization of stem cells: karyotyping, fluorescence in situ hybridization (FISH), comparative genomic hybridization, single nucleotide polymorphism (SNP) analysis, and epigenetic profiling. Some types of stem cells, particularly embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), are more prone to being genetically unstable and should be observed frequently for chromosomal changes.
Stem cells express both unique and specific combinations of transcription factors, cell surface proteins, and cytoplasmic proteins. Techniques used for stem cell analysis and characterization include flow cytometry, array-based analysis of the transcriptome, immunocytochemistry, western blots, and biomarker analysis.
Different classes and types of stem cells are characterized by different combinations of markers. In addition to confirming that stem cell lines are stable, markers can be used for screening during the reprogramming of somatic (adult) cells into induced pluripotent stem cells (iPSCs) or to follow the progression of stem cell differentiation.
Karyotyping is the examination of cellular chromosome number and morphology. Differences in appearance include size, position of centromeres, and changes in banding patterns.
Stem cells in culture can accumulate changes including chromosomal abnormalities. Alterations in the genome can lead to changes in gene expression and cellular functions of stem cells (Inzunza et al. 2004) and increase the risk of the stem cells being tumorigenic. Thus, it is crucial to monitor a culture for the any chromosomal abnormalities, particularly in stem cells intended for therapeutic use.
It is generally recommended that a stem cell line be karyotyped every 10–15 passages to ensure that chromosomal duplications, insertions, deletions, translocations, or centromere loss have not occurred.
Traditional karyotyping uses dye to stain the chromosomes of a metaphase cell in distinct banding patterns. The most common method is Giemsa staining, known as G-band karyotyping or G-banding; other methods include R-banding (reverse Giemsa staining), C-banding (constitutive heterochromatin staining), Q-banding (quinacrine staining), and T-banding (telomeric staining). Changes in banding patterns are used to identify abnormalities.
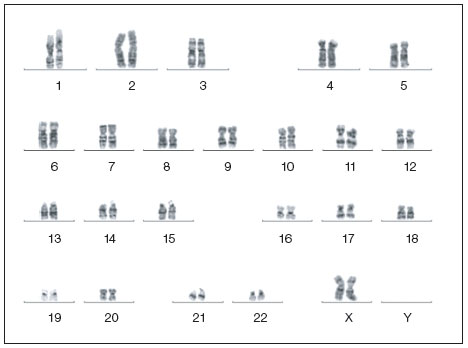
Normal karotype of a human iPSC clone. Image courtesy of Dr. Miguel Esteban.
In FISH, fluorescently labeled overlapping DNA fragments are hybridized to interphase or metaphase chromosomes. The chromosomes are formalin-fixed onto a surface and chemically denatured. Fluorescence microscopy is used to detect and analyze the chromosomes. In Fast FISH, chromosomes are denatured with high temperatures instead of chemically.
Spectral karyotyping (SKY) and multiplex fluorescence in situ hybridization (M-FISH) are commonly used forms of FISH for detection of changes in chromosomes. Both are hybridization-based combinatorial techniques using fluorescently-labeled DNA probes. Each chromosome is labeled with a different combination of fluorophores specific for that chromosome, giving a unique spectral signature for each chromosome. Chromosomes are identified by their unique signatures, and then the output is pseudo-colored to aid in visualization. The difference between SKY and M-FISH is how the fluorescent signals are collected and processed. These methods can both detect translocations within and between chromosomes more accurately than traditional karyotyping (Liehr et al. 2013); however, they cannot detect inversions or duplications and deletions of less than approximately 5 Mb.
The main disadvantage of FISH for general screening of stem cells is that probes are required for the length of all the chromosomes. FISH is most commonly used for confirmation of the findings of other techniques using probes for a specific region of a chromosome. Deletions and insertions greater than ~20 kb are detected. Fast Fish is usually used for detecting specific aneuploidies (missing or additional copies of a chromosome) that are commonly found in iPSCs and ESCs.
Comparative genomic hybridization can both determine chromosomal ploidy and detect copy number variations (CNVs), which are variable numbers of copies of segments of DNA.
In this technique, DNA samples from the test stem cell population and from a reference population are each labeled with different probes. When this technique was first developed, reference and test DNA were denatured then hybridized to metaphase chromosomes. Using microscopy and fluorescence measurements, the chromosomes were scanned for any differences in fluorescent signals between the test and reference samples (Kallioniemi et al. 1992). This technique detects and locates CNVs without requiring any knowledge of the particular sequence.
The disadvantage of using whole metaphase chromosomes is that the technique is not much more sensitive than G-banding (with a limit of detection of ~5 Mb). The adoption of techniques using microarrays with genomic clones has increased detection sensitivity and efficiency.
Comparative genomic hybridization does not detect balanced translocations, inversions, or small gains or losses. Both mosaicism in a stem cell population and the presence of repetitive regions can complicate interpretation of results when using arrays. Due to the variability in some chromosomal regions among individuals, the preferred reference is either an early passage of the stem cells or another DNA sample extracted from the source of the stem cells (for example, from host blood cells).
For analysis of known areas of CNV where sequence information is available, digital PCR is sensitive enough to detect CNV in short sequences and determine copy number with high precision.
Single-nucleotide polymorphisms (SNPs), single base-pair mutations within a region of DNA, are the most common type of genetic variation (Brookes 1999). SNPs may accumulate in a stem cell population over time and can lead to phenotypic changes that influence survival or growth. These genetic changes may result in unwanted outcomes such as loss of pluripotency or gain of tumorigenicity. SNP genotyping with high-density oligonucleotide arrays (Gunderson et al. 2005) can specifically identify a stem cell line's origin and monitor its genomic integrity. SNPs can be identified using PCR, microarrays, or DNA sequencing.
SNP arrays are used for detecting loss of heterozygosity (LOH). Normally for a chromosome pair, since the chromosomes come from two parents, there are differences between the sequences of the two chromosomes including many SNPs. When one copy of a region in a pair of chromosomes is lost, this results in LOH. Cultured hESC have been demonstrated to lose heterozygosity with passage (Närvä et al 2010). LOH can be associated with an increase in tumorigenic potential. However, LOH can also be used in research. Currently, stem cells are being used as cancer models by inducing chromosome-specific LOH.
Aneuploidy, unbalanced translocations, deletions, and duplications can be detected with SNP arrays. Inversions are not detected and low levels of mosaicism may confound interpretation. As with comparative genomic hybridization, due to variability between individuals, choice of reference material is important.
Comparative genomic hybridization and SNP detection can be combined in an array.
A stem cell's pluripotency is dictated to a large extent by its epigenetic profile (Bibikova et al. 2006). DNA methylation and histone modification regulate the accessibility of DNA to the transcriptional machinery and thus regulate gene expression (Jaenisch and Bird 2003).
Analysis of DNA methylation patterns is performed by treating DNA with bisulfite under controlled conditions such that all cytosines are converted to uracils while leaving methylated cytosines unchanged. After conversion, the DNA can be analyzed for global methylation patterns using a chip array, PCR, or DNA sequencing. Chromatin immunoprecipitation (ChIP) analyzes patterns of histone modifications by cross-linking the DNA to histones. Cross-linked chromatin is then sheared and purified using antibodies against a specific protein or histone modification. Analysis of the cross-linked regions can be performed using real-time qPCR, microarrays, or DNA sequencing. New stem cell lines can be initially characterized by examining the epigenetic state of a few key genes such as H19, Xist, Oct4, Notch 1, Dlk1/MEG3, and PWS/AS (Loring et al. 2007).
Quantitative assessment of chromatin structure in cultured cells can also be performed using specific epigenetics tools. Chromatin is digested in the presence or absence of nuclease, and then the genomic DNA is purified and quantified. Chromatin structure is assessed via real-time PCR, by comparing results against an epigenetically silenced (reference) gene to discriminate open, actively transcribed chromatin regions from closed, transcriptionally silent regions. This quantifies the impact of epigenetic events such as DNA methylation and histone modification on gene expression regulation through chromatin state changes.
Epigenetic profiling has shown that iPSCs can retain some epigenetic memory. As cells differentiate during development, there are epigenetic changes accompanying the changes in gene expression patterns. When a somatic cell is reprogrammed to become an iPSC, it has been observed in some iPSC cell lines that not all the epigenetic changes that occurred during differentiation are reversed during dedifferentiation. Newly generated iPSC lines must therefore be screened to determine whether they retain epigenetic memory.
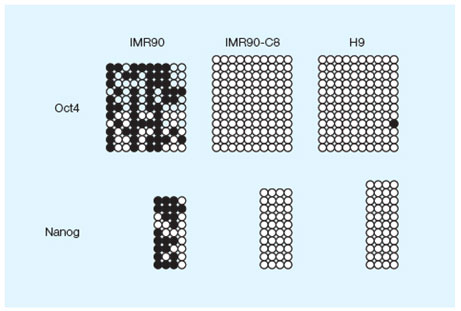
DNA methylation profile of the Oct4 and NANOG proximal promoters in three cell lines. Image courtesy of Dr. Miguel Esteban.
Flow cytometry is a commonly used technique used for the analysis of stem cells. Fluorescently tagged monoclonal antibodies can be used to distinguish specific cell populations, and cell sorting can be used to physically separate different stem cell populations. For example, when a population of somatic cells is being reprogrammed, iPSCs can be separated from cells either not reprogrammed or not fully reprogrammed. Another common use of cell sorting is during maintenance and expansion of stem cell culture. The development of mosaic populations is common in stem cells stocks. Sorting can be used to remove unwanted cell types and retain homogeneity of the population.
There are a large number of well-characterized cell surface markers that can be used both to determine cellular phenotypes and to separate mixed populations. Additionally, stem cells express a number of cell surface proteins that are used as markers of pluripotency and lineage. For example, the glycolipid antigens SSEA-3 and SSEA-4 and the keratin sulfate antigens TRA-1-60 and TRA-1-81 are commonly used antigens for identifying and sorting ESC populations (Adewumi et al. 2007).
Fixed stem cells can also be sorted based on the expression of the transcription factors such as Oct-3/4 and NANOG. As stem cells differentiate, the loss of these markers and expression of new markers can be used to track the lineages and level of differentiation of the population.
Immunocytochemistry is predominantly used to confirm the presence or absence of protein expression in stem cells. Cells (usually fixed) are incubated with antibodies targeting specific proteins prior to fluorescent imaging. In addition to providing information about the numbers of cells expressing particular proteins, information can be obtained about relative expression levels and the subcellular localization of the target proteins. Immunohistochemistry is also used to visualize stem cells in tissues, including the engraftment of stem cells after transplantation.
In experimental models of tissues and whole organisms, stem cells can be transduced to express a protein that can be easily tracked. The visualization of a protein that is not expressed in a given tissue or organism permits discrimination between transplanted stem cells and any resident stem cells. An example is green fluorescent protein (GFP), which can be tracked either directly by green fluorescence or, more commonly, using an anti-GFP antibody to take advantage of the signal amplification afforded by antibody sandwich assays.
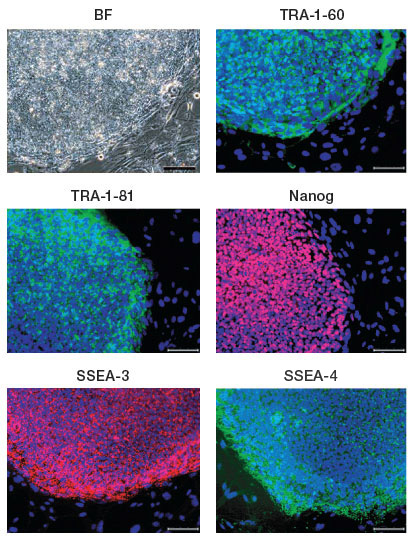
iPSC clones generated from the human fibroblast cell line IMR90 expressing the human ESC surface markers TRA1-60, TRA-1-81, SSEA-3, SSEA-4, and the transcription factor NANOG; nuclei are stained blue. Image courtesy of Dr. Miguel Esteban.
Reverse transcription quantitative PCR (RT-qPCR) and RT-digital PCR provide rapid, sensitive, and quantitative methods for monitoring the gene expression profile of any cell population (see the Real-Time PCR and Digital PCR pages for comprehensive descriptions of these methods).

RT-PCR analysis of endogenous OCT4, SOX2, and NANOG and markers for the three germ layers in embryoid bodies in three different cell lines. Image courtesy of Dr. Miguel Esteban.
The levels of transcription factors such as Oct4, NANOG, SOX2, and other synergistic factors determine whether pluripotency is maintained (Li 2010). Multiplexed RT-qPCR assays and panels provide an efficient way to screen and quantify transcription factors, kinases, and other molecules involved in both maintaining pluripotency and in differentiation. The ability to do multiplex transcriptome analysis increases the sensitivity of determining the status of cells.
Digital PCR can increase the accuracy of measurements with lower amounts of input nucleic acid. Droplet Digital™ PCR allows for absolute quantification of transcripts and detection of rare sequences. In addition to being useful for transcriptome analysis, digital PCR can accurately detect CNVs and SNPs.
In studying the mechanism of stem cell differentiation, western blotting is useful for determining the success of transfection experiments. When a gene is either introduced into the cell or knocked down using antisense RNA or RNAi molecules, detection and quantitation of the presence and level of protein expression by western blotting is often coupled with measurements of transcript levels by RT-qPCR. Importantly, western blots can be used to study the effects of transfection on downstream protein expression.

Ascorbic acid (vitamin C; Vc) reduces senescence in three mouse embryonic fibroblasts (MEF) cell lines transduced with the SKO transcription factors Sox2, Klf4, and Oct4. The western blot demonstrates that p21 expression is reduced after Vc treatment (top row) vs. the tubulin loading control (bottom row). Images courtesy of Dr Miguel Esteban.
Regulation of biological processes can be controlled by protein interactions, relative protein levels, and posttranslational protein modification. Not only can the presence or absence of proteins or signaling peptides be assessed, but relative levels can be determined. This technique can be used to evaluate pluripotency, lineages, and differentiation.
The ability to multiplex allows for fine analysis of stem cells and differentiation pathways. Biological reactions and signaling networks can be monitored in real time. The use of bead-based multiplex immunoassays allows for simultaneous monitoring of protein, cytokines, and chemokines and can also assess the phosphorylation states of many proteins involved in signal transduction.
Both embryoid body (EB) and teratoma formation can be used as indicators of stemness when analyzing the pluripotency of stem cells. Both will differentiate and contain cells from all three germ layers, demonstrating pluripotency. EBs will develop spontaneously in ESC or iPSC cultures upon withdrawal of factors that maintain pluripotency, when cultures become very dense, or when suspended in media that promotes formation of EBs.
Teratomas are tumors containing differentiated and undifferentiated cells from all three germ layers. Generation of teratomas is a method for functional analysis of pluripotent stem cells in vivo. Stem cells are injected into immunocompromised mice. The resulting teratomas are analyzed for the presence of cells from all three germ layers.
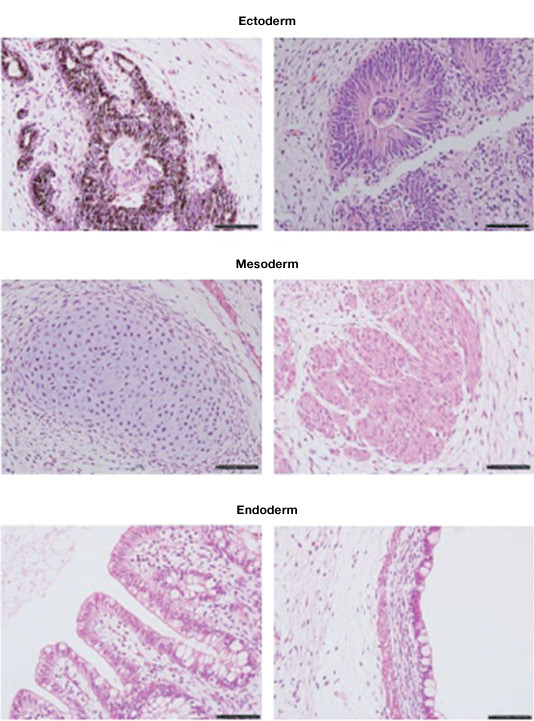
Teratomas composed of tissues derived from the three germ layers produced after injection of a stem cell line into immunosuppressed mice. Image courtesy of Dr. Miguel Esteban.
Adewumi O et al. (2007). Characterization of human embryonic stem cell lines by the international stem cell initiative. Nat Biotechnol 25, 803–816. PMID: 17572666
Bibikova M et al. (2006). Human embryonic stem cells have a unique epigenetic signature. Genome Res 16, 1075–1083. PMID: 16899657
Brookes AJ (1999). The essence of SNPs. Gene 234, 177–186. PMID: 10395891
Gunderson KL et al. (2005). A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet 37, 549–554. PMID: 15838508
Inzunza J et al. (2004). Comparative genomic hybridization and karyotyping of human embryonic stem cells reveals the occurrence of an isodicentric X chromosome after long-term cultivation. Mol Hum Reprod 10, 461–466. PMID: 15044603
Jaenisch R and Bird A (2003). Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33 Suppl, 245–254. PMID: 12610534
Kallioniemi A et al. (1992). Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258, 818–821. PMID: 1359641
Li YQ (2010). Master stem cell transcription factors and signaling regulation. Cell Reprogram 12, 3–13. PMID: 20132009
Liehr T et al. (2004). Multicolor fish probe sets and their applications. Histol Histopathol 19, 229–237. PMID: 14702191
Loring et al. (2007). Human Stem Cell Manual: A Laboratory Guide (Academic Press).
Myers JD (1989). Development and application of immunocytochemical staining techniques: A review. Diag Cytopathol 5, 318–330. PMID: 2477206
Närvä E et al. (2010). High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol 28, 371-377. PMID: 20351689
