- Targeted Genome Editing
- Targeted Genome Editing Applications
Targeted genome editing is having a major impact on many applications including drug discovery. The low cost, fast turnaround, and adaptability of editing with CRISPR/Cas9 constructs permits more detailed analysis than previous editing and interference techniques. In addition to modulating gene expression, analysis and screening is being rapidly expanded to previously unexplored noncoding regions of the genome to look for regulatory sequence and druggable targets. The ability to rapidly generate model systems that are more physiologically relevant or specific to individual testing paradigms increases the likelihood that findings in a model system will translate to positive clinical trials.
Page Contents
CRISPR/Cas9 is a very effective tool for forward genetic screening. In forward genetic screening, mutations are used to identify genes and noncoding regions that are involved in a phenotype, whereas in reverse genetic screening (the most common type of screening using nonediting techniques), a specific gene is mutated to determine the effects on phenotype. Forward genetic screens can provide information about previously unidentified genes, hitherto unknown functions of known genes, regulatory sequences, and roles of regulatory RNAs such as long noncoding RNA (lncRNA) and other RNAs not involved in transcription.
CRISPR/Cas9 libraries are frequently used to carry out large-scale genomic screening. Tiling libraries, which contain overlapping RNA sequences covering a region of interest, permit screening of both coding and noncoding sequences. Besides directly assessing alterations in phenotype, this type of screening can be used to study the regulation of the effects of drugs. For example, a CRISPR/Cas9 screen of the noncoding regions around three genes associated with resistance to chemotherapy found mutations affecting the expression of these genes. Further mutations in sequences around one of these genes was found to confer resistance to vemurafenib, a drug used for treating late-stage melanoma (Shalem et al., 2014). In another approach, the error-prone nature of repair by non-homologous end joining (NHEJ) with the CRISPR/Cas9 system was used to identify gain-of-function and drug-resistant variants of the MAPK, MEK1 (Donovan et al. 2017).
An extremely active area of research is targeted genomic editing in stem cells. In addition to studies of the physiology and functioning of different types of progenitor cells, gene edits in pluripotent stem cells are being used to develop models of diseases. These disease-specific stem cells are being used to increase understanding of diseases and for rapid, large-scale screening of drugs and other therapeutic modalities for disease treatment. Recent advances in techniques such as 3D printing of cells and development of scaffolds is permitting the use of 3D tissue models for screening.
One of the promises of stem cell therapy is the ability to use a patient’s own stem cells (removed from the patient or from previously banked cord blood) to correct a defect and then inject these corrected cells therapeutically into the patient. Targeted gene editing techniques are providing a fast, efficient method for correcting mutations or creating other desired changes in sequence.
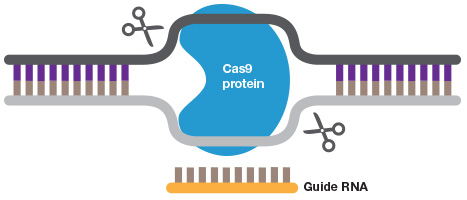
Gene editing. A short guide RNA can be used to target Cas9 to a sequence of interest. Cas9 introduces a double strand DNA break to allow highly efficient gene editing.
A gene drive is a mechanism for the introduction of a trait into the germline of an organism, which then actively propagates in a population through breeding. An example is the microinjection of a CRISPR/Cas9 construct that creates resistance to the malaria parasite Plasmodium falciparum into mosquito embryos (Gantz et al, 2015). When adult mosquitoes with the CRISPR/Cas9 construct integrated into their DNA breed with wild-type mosquitoes, there will be an allele containing the CRISPR/Cas9 construct and a wild-type allele in the offspring. The CRISPR/Cas9 construct will edit the allele derived from the wild-type parent, making all offspring homozygous for parasite resistance. Release of mosquitos carrying the altered germline into the wild will result in a rapidly increasing proportion of mosquitos becoming resistant to the parasite with each subsequent generation.
Constructs with inactivated Cas9 endonuclease activity (dCas9) are being used to activate or repress gene expression. Since Cas9 nuclease activity is independent of binding specificity, CRISPR/dCas9 constructs with mutations in the active site of the nuclease still retain targeted binding. Fusing a transcriptional activator or repressor to a CRISPR/dCas9 construct that is targeted to a transcriptional start site or other regulatory sequence can be used to either activate or upregulate (CRISPRa) or downregulate (CRISPRi) gene activity.
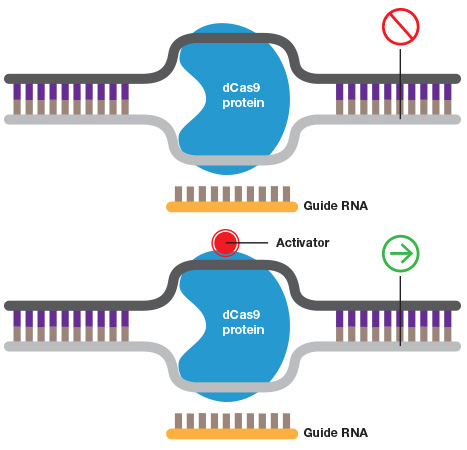
Turning genes on and off. A guide RNA can also be used to target catalytically inactive Cas9 (dCas9) to a promoter to inhibit transcription by blocking transcriptional machinery from binding or to activate transcription when dCAs9 is fused to an activator.
Before the use of CRISPR/dCas9, the main technique used for downregulation of transcription was RNAi. CRISPRi is at least as efficient as RNAi in repressing transcription. Upregulation of genes previously required cloning of construct containing a promoter and open reading frame and then introducing this construct in cells. The advantage of CRISPRa is that the endogenous gene is targeted and upregulated rather than the increase in transcripts coming from a construct somewhere in the cell.
Further, with the addition of some form of a "switch" mechanism, the activation or repression can be temporally controlled. For example, a light-inducible system was developed, using a complex containing CRISPR/dCas9 and eGFP fused to two proteins that can dimerize, thus activating a transactivation domain. This system provides an easy-to-use mechanism for light-activated transcription (Polstein and Gersbach, 2015).
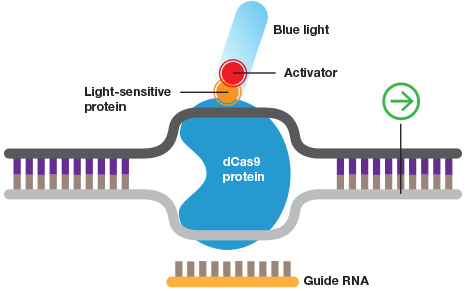
Inducible CRISPR. Wild-type Cas9 or dCas9 can be fused to switches that are controlled by light or a specific chemical.
In addition to changes in the DNA sequence, modifications of nucleotides, such as cytosine methylation and histone modification, can also cause heritable changes in gene expression. Using constructs with CRISPR/dCas9 fused to enzymes such as demethylases and acetyltransferases specific modifications can be targeted and altered. This permits analysis of the role of individual modifications and the study of their effects on gene expression and modulation.
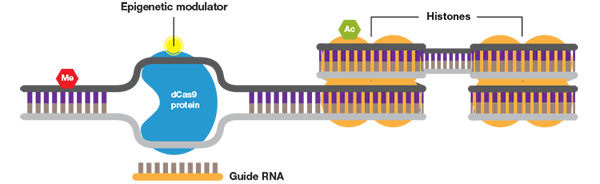
CRISP epigenetics. When fused to a DNA methylase or histone acetylase, the CRISPR system can be used to study the effects of specific DNA and histone modifications.
Targeted genome editing is having a major impact on drug discovery. The low cost and fast turnaround of editing with CRISPR/Cas9 constructs permits more detailed analysis than previous editing and interference techniques for genes or regulatory regions. Analysis and screening is being rapidly expanded to previously unexplored regions of the genome to look for new drug targets.
An example of a major use of targeted editing is to generate cell lines that mimic diseases. One advantage of the CRISPR/Cas9 system, with its low cost and high speed, is that multiple models can be developed for the same disease, including more physiologically relevant models in stem cells. If a candidate lead drug is effective in all models, then this will increase the probability that a treatment will be effective in clinical trials. Since most candidate drugs fail after the initial testing, any increase in the percentage of candidate lead drugs that make it to market could provide significant cost savings.
A current avenue of research is the use of CRISPR/Cas9 as a direct or indirect therapeutic agent. One area of research involves the development of systems where CRISPR/Cas9 can be used as antimicrobials. A sequence targeting a microbial genome could be specifically directed to inactivate a sequence conferring antibiotic resistance or a sequence that is necessary for virulence or even viability. This should provide much faster identification and time to market for new antibiotics. Another example of an antimicrobial strategy could be treating immunosuppressed individuals after organ transplantation to prevent replication of viruses, such as herpes virus.
A delivery system for targeted genome editing that is being investigated is the use of modified immune cells for personalized treatments. After removal of immune cells from a patient, the desired trait is introduced via targeted genome editing and after expansion the immune cells are reintroduced to the patient. These cells could be engineered to attack targets, such as viruses or cancer cells.
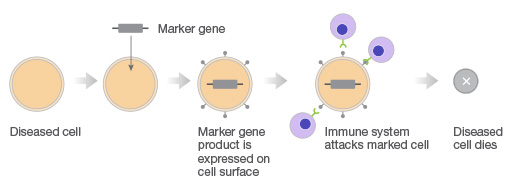
Gene therapy with CRISPR. Gene therapy approach using transgenes as suicide genes that kill target cells or as molecular beacons that mark specific cells for destruction.
Another approach developed by Sangamo Therapeutics is to use zinc finger nucleases (ZFNs) to introduce a mutation in a gene that confers resistance to infection by HIV. Cells from a patient's immune system T-cells, and hematopoietic stem cells (which give rise to T-cells) are edited to knock out the CCR5 gene with the same mutation that is found in "long-term non-progressors" with HIV infections. This treatment is currently in clinical trials. The Sangamo team uses Droplet Digital PCR to verify gene edits and confirm the very low to no residual viral load in treated patients, which are not reliably detectible with conventional qPCR assays.
Cell and animal models have been used to demonstrate the utility of targeted gene editing to correct mutations in monogenic diseases by permanently altering the germline. CRISPR knock-in and knockout rodents are now available commercially and the CRISPR/Cas9 system is rapidly replacing "traditional" methods of creating animal models with germline alterations. However, there are still many technical and ethical issues that must be addressed before targeted genomic editing is used for gene therapy in humans.
There are many different CRISPR/Cas9 vectors available for targeted genomic editing, most commonly plasmid or viral. Vectors are usually engineered for a specific type of editing. Many of these vectors include a sequence that will provide the ability to select for cells containing edits. The most common are GFP and antibiotic resistance. GFP fluorescence can be used for cell sorting, while antibiotic resistance will eliminate nonedited cells. Introduction of constructs into cells is performed using standard transfection or transduction methods.
The methods used for screening, such as identification of edits and functional assays, depend on the application. Screening of base mismatches and small indels (insertions and deletions) can be accomplished using a surveyor nuclease assay. After PCR amplification of both wild type and edited genomic DNA, hybrid heteroduplexes are digested with a surveyor nuclease (mismatch-specific endonuclease) followed by analysis of the fragments. Sequence analysis can provide the location and surrounding sequence information for predicted sites of off-target editing.
More recently, digital PCR is providing a method that is faster, easier, and more sensitive for detection and quantification of genome edits (see Bulletin 6712: Ultra-Sensitive Quantification of Genome Editing Events Using Droplet Digital PCR). Digital PCR can be used directly on isolated DNA, obviating the need for manipulation and digestion of genomic DNA. Though digital PCR and surveyor assays can be used to detect any predicted off-target events, there can often be edits that were not predicted. Next-generation sequencing (NGS) analysis of the entire genome can identify all edits. Digital PCR Library Quantification Kits provide quantification and balancing of sequencing libraries to ensure that all edits are detected.
A typical targeted gene editing experiment involves rendering a gene nonfunctional (gene knock-out) by replacing it with a gene expressing a reporter protein, e.g., a fluorescent protein, which can be used both to confirm knock-out and to select for edited cells. Different methods and products may be used in each experiment stage.
For more information on gene editing workflows and tools, see CRISPR Gene Editing Workflow.
Sangamo Therapeutics targeted gene editing therapies in clinical trials: Clinical Trials.gov
Donovan KF et al. (2017) Creation of novel protein variants with CRISPR/Cas9-mediated mutagenesis: Turning a screening by-product into a discovery tool. PLOS One 12, e0170445. PMID: 28118392
Gantz VM (2015) Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 112, E6736-43. PMID: 26598698
Polstein LR and Gersbach CA (2015) A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11, 198-200. PMID: 25664691
Shalem O et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84-7. PMID: 24336571
Further Reading
Quinlan A (2017). Erasing disease: How gene editing is changing genetic therapies., accessed April 4, 2017
Quinlan A (2016). CRISPR: Changing the pace of BioPharma R&D., accessed April 4, 2017
Quinlan A (2016). From megaTALs to CRISPR: The many ways to edit a gene., accessed April 4, 2017
