On This Page |
Bacterial Transformation | pGLO Plasmid | Gene Regulation | Green Fluorescent Protein | Other Resources | pGLO Bacterial Transformation and GFP Kits |
Green fluorescent protein (GFP) is a protein that glows with a bright green fluorescence under ultraviolet light. First isolated from the marine jellyfish Aequorea victoria, the gene encoding GFP is used in cellular and molecular biology as a reporter to detect gene expression in transgenic organisms.
Bio-Rad Explorer pGLO Plasmid and GFP Kits use the pGLO plasmid, which contains the GFP gene, to enable hands-on learning about the central dogma, gene expression and regulation, bacterial transformation, protein separation, and the biomanufacturing process.
Bacterial Transformation
With pGLO bacterial transformation, students learn about genetic engineering as they transform a non-virulent laboratory strain of Escherichia coli (E. coli) with the pGLO plasmid. The procedure involves the CaCl2/heat shock method, which is a standard technique used in many research and biomanufacturing laboratories.

pGLO Bacterial Transformation Powerpoint (PPT 1.5 MB)
This editable presentation provides an overview of bacterial transformation with the pGLO plasmid.

How To Perform a Bacterial Transformation Video
This video demonstrates how to transform bacteria using the Bio-Rad Explorer pGLO Bacterial Transformation Kit.
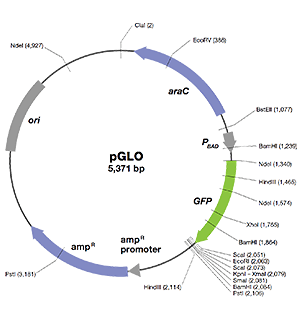
pGLO Plasmid
Bio-Rad’s pGLO plasmid contains DNA sequences that enable its replication and expression of the fluorescent trait (phenotype) in bacteria following transformation. The essential sequences include the following:
- GFP — jellyfish gene that encodes green fluorescent protein (GFP)
- ori — origin of pGLO plasmid DNA replication (essential for making more copies of the plasmid)
- bla — gene that encodes β-lactamase, an enzyme that breaks down the antibiotic ampicillin; transformants expressing the bla gene can be selected by placing ampicillin in the growth medium
- pBAD promoter — binds AraC-arabinose and promotes RNA polymerase binding and transcription of the GFP gene
- araC — gene that encodes the regulatory protein that binds to the pBAD promoter; only when arabinose binds to the AraC protein is the production of GFP switched on
- Multiple cloning site — a region containing restriction sites (NdeI, HindIII, EcoRI, etc.), sequences that permit the insertion or deletion of a gene of interest
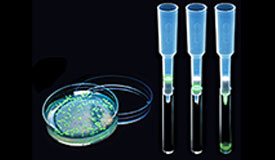
Bacteria transformed with the pGLO plasmid are selected by ampicillin resistance and when induced to express GFP, they glow fluorescent green under UV light!
Gene Regulation
Gene expression is carefully regulated to allow organisms to adapt to differing conditions and prevent wasteful production of proteins. Regulation often occurs at the level of transcription from DNA into RNA, specifically at the promoter, where RNA polymerase binds the DNA and begins transcription of the gene.
In bacteria, groups of related genes are often clustered together and transcribed into RNA from one promoter. These clusters of genes controlled by a single promoter are called operons. The bacterial genes encoding the enzymes needed to metabolize the simple sugar arabinose are a perfect example. Three genes that encode the digestive enzymes involved in breaking down arabinose (araB, araA, and araD) are clustered together in arabinose operon 3, and all depend on initiation of transcription from a single promoter, pBAD. Transcription requires the simultaneous presence of RNA polymerase, a DNA-binding protein called AraC, and arabinose.
- When arabinose is absent, the AraC protein binds to the DNA at the binding site for RNA polymerase, preventing transcription of the digestive enzymes
- When arabinose is present, it interacts with AraC, causing AraC to change shape, allowing RNA polymerase to bind the promoter; araB, araA, and araD are then expressed and can do their job to break down arabinose until the arabinose runs out
The pGLO plasmid contains both the promoter (pBAD) and araC gene, but araB, araA, and araD have been replaced by the single gene that codes for GFP, which serves as a reporter gene. In the presence of arabinose, the AraC protein promotes the binding of RNA polymerase to the promoter, which causes transcription of the GFP gene into messenger RNA (mRNA), followed by the translation of this mRNA into GFP. This process is called gene expression.
As they produce more and more protein, the cells expressing GFP fluoresce a brilliant green. In the absence of arabinose, however, AraC no longer facilitates the binding of RNA polymerase, and the GFP gene is not expressed, and bacterial colonies have a wild-type (natural) phenotype — white colonies with no fluorescence.
This is an excellent example of the central dogma of molecular biology in action: DNA > RNA > protein > trait.
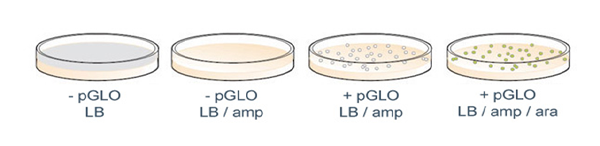
Results of a pGLO bacterial transformation experiment. Controls that were incubated with no plasmid (-pGLO) grow as a lawn in the absence of ampicillin (LB plate) and do not grow at all in the presence of ampicillin (LB/amp). Transformants grown on ampicillin (LB/amp) grow as colonies but do not show GFP fluorescence; those grown in the presence of both amp and ara (LB/amp/ara) do glow green under UV light.
Green Fluorescent Protein
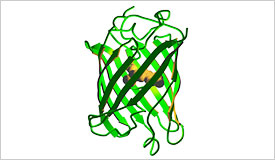
GFP has a barrel structure surrounding a central alpha helix that contains the fluorophore. It can be used as an example for discussions of protein secondary structure, parallel and anti-parallel beta sheets, and the use of genes and proteins in biotechnology. For more information, visit our partner 3-D Molecular Designs.
GFP Purification — Electrophoresis and Chromatography (PPT 9.63 MB)
The GFP expressed from the pGLO plasmid illustrates the central doctrine of biology, from the transformation of DNA to the expression of a protein to the visualization of a trait. The bacterial proteome contains thousands of proteins, but only the cloned GFP glows!
In its native environment, GFP fluoresces in the deep sea jellyfish, Aequorea victoria. Incredibly, GFP retains its fluorescent properties when cloned and expressed in E. coli and even when isolated from E. coli and separated on polyacrylamide gels or by chromatography. These amazing properties of GFP allow students to visualize the phenotypic properties of a protein and identify the single protein “band” responsible for the trait. These extensions link two of the most commonly used techniques in biotechnology labs: transformation and protein purification.
Purification of a protein depends on a its chemical or physical properties, such as molecular weight, electrical charge, or solubility. GFP can be separated from others by its size using electrophoresis, and it is extremely hydrophobic, which enables its purification using hydrophobic interaction chromatography (HIC). When placed in a buffer containing a high concentration of salt, the HIC matrix selectively binds hydrophobic GFP molecules while allowing the bacterial proteins to pass right through the column. Then, simply lowering the salt concentration of the buffer causes GFP to elute from the column in a purer form.
Students can explore these separation techniques by growing transformed bacteria in liquid culture to grow overnight, then lysing the cells to release their contents. The unique fluorescent property of GFP allows real-time monitoring of extraction and purification, modeling key processes used in biotechnology to produce and purify designer proteins with commercial or research value.
Other Resources
Bring Inquiry Into Your Classroom with the pGLO Plasmid (PPT 9.06 MB)
Use pGLO Bacterial Transformation to illustrate the science and engineering practices described in the NGSS framework.

YouTube pGLO Bacterial Transformation Playlist
Use these short, instructional videos to enrich lessons about bacteria, bacterial transformation, and the green fluorescent protein (GFP).
Case Studies
Student-facing extensions that are also useful for AP exam prep.

Case Study: A Role for Bacterial Transformation in Controlling Malaria Transmission (PDF 3.3 MB)
Case Study: Hacking the Gut Microbiome (PDF 1.5 MB)
pGLO Bacterial Transformation and GFP Kits
pGLO Lab Kits utilize Bio-Rad’s pGLO plasmid, which encodes a green fluorescent protein (GFP), to enable instructors to give students a hands-on introduction to transformation, cloning, protein chromatography, and electrophoresis techniques.
pGLO Bacterial Transformation Kit
Use bacterial transformation with an inducible promoter to make glowing
E. coli.
pGLO Bacterial Transformation Kit for General Biology
Apply a three-dimensional approach to the classic pGLO bacterial transformation activity.
pGLO Transformation and Inquiry Kit for AP Biology
Investigate the functional elements of pGLO bacterial transformation, including heat shock, antibiotic selection, promoters, and satellite colony formation.
pGLO SDS-PAGE Extension
Use protein electrophoresis to view the expression of proteins in your pGLO bacteria.
Green Fluorescent Protein Chromatography Kit
Use chromatography to purify glowing green fluorescent protein from your pGLO bacteria.
Secrets of the Rainforest Kit
Simulate the drug discovery process in your classroom with a glowing protein.