Certain real-time PCR instruments require an internal reference dye for fluorescent signal normalization and correction of well-to-well optical variations. Variation in fluorescence can result from instrument design. ROX passive reference dye is an inert additive that provides a constant fluorescent signal for sample normalization throughout the real-time qPCR assay.
Related Topics: qPCR Instrumentation, qPCR/Real-Time PCR Reagents, and qPCR Assay Design and Optimization.
Page Contents
A Cq (quantification cycle) value is an exponential number derived from the intersection of the threshold and a given amplification curve and is a relative concentration measurement of the target presence in the qPCR reaction. Several factors independent of the sample influence the Cq value — the optical system, PCR supermix, threshold and baseline settings, and the presence or absence of a passive reference dye such as ROX. Before explaining how a passive reference dye like ROX impacts Cq values and qPCR data, it is important to understand how optical real-time PCR detection systems work.
The optical design of most real-time PCR systems, using a lamp or other single stationary light source, creates illumination and detection light paths that vary for each well of the thermal cycler block. The variation in light path lengths produces different absolute fluorescence measurements for wells that contain the same concentration of a reporter fluorophore. A well with a shorter light path, typically located in the middle of the block, will have a higher fluorescence reading compared to a well with a longer light path, typically located on the perimeter of the block (Figure 1).
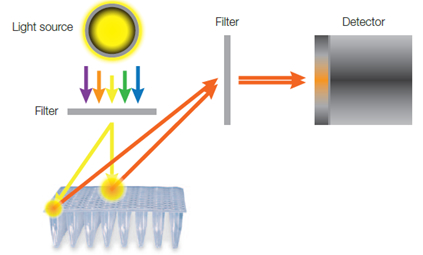
Fig. 1. Light path differences between wells of a lamp-based real-time PCR system. The well in the center of the block has a shorter light path and, subsequently, emits a higher fluorescence signal compared to the well on the perimeter, which has a longer light path.
The differences in absolute fluorescence levels do not impact real-time PCR results because results are determined by measuring the change in fluorescence of a reporter measured over the course of the reaction. For real-time PCR data processing, the sample fluorescence for each well is first baseline subtracted and then the quantification cycle (Cq) value is determined as a cycle number at which fluorescence has increased above background.
Some lamp- and laser-based real-time PCR systems utilize an internal reference dye to normalize well-to-well fluorescence signal differences resulting from optical path length variations. Normalizing the fluorescence intensity is accomplished in real-time PCR software by dividing the emission intensity of the reporter dye by the emission intensity of a reference dye. In contrast to these real-time PCR systems, Bio-Rad real-time PCR detection systems utilize solid-state optical technology with long-lasting light-emitting diodes (LEDs) for fluorescence excitation and photodiodes for detection. At every position and on every scan, the optics shuttle is reproducibly centered above each well, so the light path is always identical for all wells.
ROX (carboxy-X-rhodamine) is used as a passive reference dye in ROX-dependent real-time PCR systems to normalize for differences in fluorescence levels that can occur due mainly to optical path variations among wells. ROX is not involved in the PCR reaction, and ROX fluorescence levels have no relationship to the quantity of DNA in each well, so the addition of this fluorophore to a supermix provides a constant fluorescent signal during the amplification cycles. The constancy of ROX fluorescence relative to reporter fluorescence signal is used for well-to-well fluorescence normalization.
Different types of real-time PCR systems that require a passive reference standard have different optimal concentrations of ROX, mainly due to the different optical configurations of each system, particularly the type of excitation source and optics used. For some PCR master mixes, the user must determine the correct ROX concentration to optimize real-time PCR results; however, using Bio-Rad universal real-time PCR supermixes, there is no guesswork — our universal reagents can be used on any real-time PCR system.
To normalize real-time PCR data, the fluorescence emission intensity of the reporter dye (for example, SYBR® Green) is divided by the fluorescence emission intensity of the ROX passive reference dye. This ratio is the normalized reporter intensity, or Rn. Table 1 shows the calculation of Rn for two example wells highlighted in Figure 1. To further normalize the data, Rn+ and Rn- are calculated. Rn+ is the Rn of reactions that contain all components of the reaction including the template. Rn- is the baseline fluorescence, which is derived from the early cycles before there is a significant increase in reporter fluorescence. To determine the reporter fluorescence above baseline, ΔRn is calculated as (Rn+) – (Rn-) to increase the accuracy of the fluorescence measurements.
Table 1. Calculation of Rn for two wells at different positions. Fluorescence variation is corrected by normalizing the fluorescent reporter signal to the fluorescent signal of a passive reference dye. RFU, relative fluorescence units.
| Fluorescence Intensity, RFU | |||
| Well | Reporter | Passive Reference | Rn |
| Center | 1,500 | 750 | 2 |
| Perimeter | 1,000 | 500 | 2 |
To determine the initial concentration of input DNA, a threshold is chosen from the ΔRn amplification plot that represents a detectable signal from product, and is on the exponential part of the curve. Where the curve crosses the threshold is known as the quantification cycle, or Cq.
Figure 2 demonstrates the changes that occur with data normalization. The curves in the unnormalized data without ROX (A) are affected by their relative well positions. The amplification traces are not uniform, as ROX correction has not been applied. The Rn values are then baseline corrected by calculating ΔRn (B).
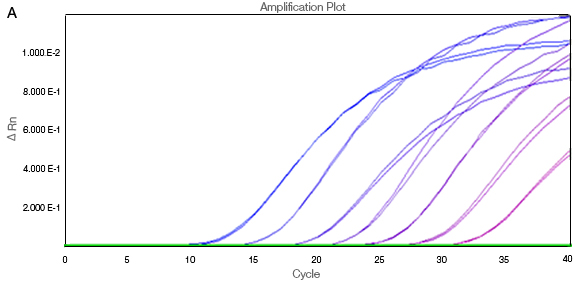
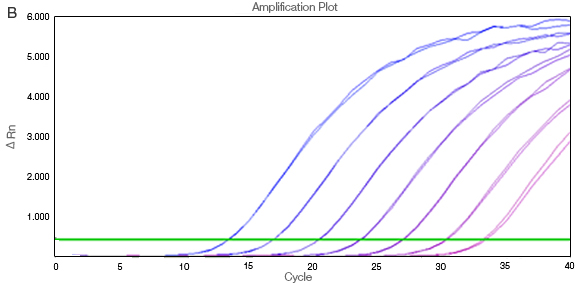
Fig. 2. Passive reference dye correction for differences in optical paths between wells. A, ΔRn vs. cycle without ROX normalization; B, baseline-corrected ΔRn values with ROX.
Does the Use of ROX Compensate for Differences in Well Volume?
Normalization of real-time PCR data using ROX or other passive reference dye also reduces, to a small extent, the effects on data of pipetting or other volume errors.
Figure 3 displays the results of a trial in which the effects of pipetting error were mimicked by varying the reaction volumes from 12 to 17 µl (n = 8) while holding the amount of input template constant at 25 ng cDNA. ROX normalization decreased the standard deviation. Additionally, the Cq values were decreased by as much as 1.6 cycles for a 15 µl reaction (Jordan and Kurtz 2010).
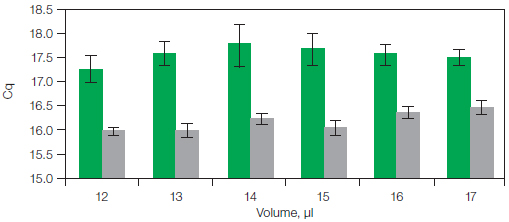
Fig. 3. Results on the ABI 7500 system for reactions with the same amount of input template and increasing final reaction volumes. Average Cq values were measured for eight replicate reactions with identical input template amount (25 ng cDNA) and variable reaction volumes (12–17 μl). ROX normalization ( ) lowered the standard deviations of Cq values for each replicate reaction set compared to Cq values for the identical reaction sets without ROX normalization (
) lowered the standard deviations of Cq values for each replicate reaction set compared to Cq values for the identical reaction sets without ROX normalization ( ). Cq, quantification cycle.
). Cq, quantification cycle.
The data in Figure 3 were obtained using an Applied Biosystems 7500 system, which is a lamp-based system. Bio-Rad real-time PCR systems use an optical shuttle, resulting in an identical light path for each well. The same protocol for changes in sample volume was repeated using the CFX96™ real-time PCR system. The results, shown in Figure 4, demonstrate that the optical shuttle gives more precise data with lower standard deviations. However, for both instruments, although the change in volume was as much as 40% between reactions, the volume change had no significant impact on the results, due to ROX normalization.
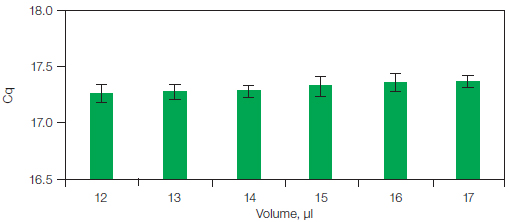
Fig. 4. Results on the CFX96 system for reactions with the same amount of input template and increasing final reaction volumes. Average Cq values were measured for eight replicate reactions with identical input template amount (25 ng cDNA) and variable reaction volumes (12–17 µl). Cq, quantification cycle.
The addition of ROX to real-time PCR reactions can increase accuracy and reproducibility on ROX-dependent real-time PCR systems. ROX normalization can be used to compensate for spurious interwell variations in fluorescence intensity due to differences in the length of optical path, minor changes in well volume due to pipetting or other errors, and some effects of bubbles in wells. Although Bio-Rad real-time PCR detection systems do not require ROX for internal normalization, our universal real-time PCR supermixes perform equally well in both ROX-dependent and ROX-independent instruments.
Jordan L and Kurtz R (2010). Optical design of CFX96™ real-time PCR detection system eliminates the requirement of a passive reference dye. Bio-Rad Bulletin 6047.
Videos
Watch this short video to find out what factors affect real-time PCR results: advanced enzymes and buffer formulations provide superior inhibitor tolerance, increased processivity, and greater speed without affecting qPCR sensitivity, efficiency, or reproducibility. Learn how to compare your current master mix to a Bio-Rad supermix.
This video explains the principles behind the polymerase chain reaction (PCR) and quantitative real-time PCR (qPCR), a technique using fluorescent detection of PCR products in real time to track exponential DNA amplification by PCR coupled with analytic methods to calculate the initial number of copies of a target DNA sequence with high sensitivity and accuracy.
FAQ
 FAQ
FAQ FAQ
FAQ FAQ
FAQ FAQ
FAQ FAQ
FAQ FAQ
FAQ FAQ
FAQ FAQ
FAQ FAQ
FAQ FAQ
FAQ FAQ
FAQ