
Proteomics is the large-scale study of protein characteristics and functions. The goal of proteomics research is to obtain an integrated view of normal and abnormal cellular/organismal processes at the level of their constituent proteins (for example, in terms of protein abundance, posttranslational modifications, protein-protein interactions, and their regulatory networks).
Proteomics can be divided into two main subcategories: protein profiling (the discovery/identification of specific targets and markers) and functional proteomics (the definition of structure, interactions, and function). 2-D PAGE is particularly well suited to protein profiling studies. It also lends itself well to targeted (functional) proteomic research, where the expression/modification of particular proteins is followed during systematic treatment regimens or alteration in growth conditions.
Protein profiling involves comparison of 2-D gels to understand various biological processes by determining the presence or absence, up or down regulation, and modification states of proteins. Profiling thus serves as a method of discovering putatively causal correlations between protein abundance or modification states and biological processes of interest. Functional proteomics aims to test specific predictions via a more detailed analysis of proteins' structures, roles, cellular locations, and interactions with other proteins.
Page Contents

A typical proteomics experiment (such as protein expression profiling) can be broken down into a series of steps. First, the experiment is designed so that the key parameters of the study have been vetted, transcribed, and reviewed. Second, extraction, fractionation, and solubilization of proteins from a cell line, tissue, or organism is carried out. This could also include processing for partial or complete depletion of high-abundance proteins for increased sensitivity of downstream analyses. Labeling of proteins with differential dyes may also be carried out to delineate effects of various treatments. In the third step, gel-based separation of proteins in mixtures is carried out in one dimension alone (SDS-PAGE) or two successive dimensions for more complex mixtures (IEF followed by SDS-PAGE). This is followed by gel staining or imaging analysis to allow for visualization, isolation and relative quantitation of proteins. Protein spots/bands of interest are excised, digested with trypsin, and identified by mass spectrometry. Additional, functional characterization of identified proteins may be done by various means.
The initial goal of most proteomics projects is to identify and determine differential abundance of proteins or posttranslational modifications (PTMs) between samples. Once a list of differentially affected proteins or their modified variants has been established, the subsequent step is to perform a detailed analysis of individual proteins of interest. This may require their expression and purification, structural characterization, assessment of biochemical activity, identification of interacting partners, or production of antibodies for additional studies. Because these analyses are time consuming and costly, accurate identification of differentially expressed proteins is critical. Unlike shotgun proteomics experiments, 2-D gel electrophoresis followed by mass spectrometry provides direct visual confirmation of changes in protein/PTM abundance, thus providing early justification for downstream analytical steps. A few additional applications of profiling are discussed below.
- Biomarker discovery — proteomic profiling allows for biomarker discovery based on the differences in protein expression levels between samples that have been analyzed on a broad scale; that is, normal vs. disease model study, protein expression knockdown by siRNA or other conditions, etc.
- Product characterization — profiling purified proteins, antibodies or samples during drug development as part of product characterization to detect batch-to-batch differences. Compared to SDS-PAGE, which provides only molecular weight information of purified proteins, 2-D electrophoresis additionally enables confirmation of uniform charge states between multiple batches
- Protein purification — this method can be used to isolate contaminants during protein purification. Profiling samples at each step in the process can assess the purity of the end product
- Host cell protein analysis — this streamlined HCP workflow allows the evaluation of complex antibodies when screening biopharmaceuticals (biologics) according to regulatory guidelines

2-D gels of total protein extracted from HeLa cells at 48 h after transfection with eGFP siRNA (control A) or actin siRNA ACT9 (B). Enlarged images of selected areas (red rectangles, C–H) show five differentially expressed protein spots (1–5). Spot 6 showed similar intensities in both gels and served as a reference.
A critical modulator of biological activity in the proteome is posttranslational modifications, or PTMs. 2-D electrophoresis can be used effectively to study PTMs. No other technique separates charge and size isomers of polypeptides as well as 2-D electrophoresis. Hundreds to thousands of polypeptides can be resolved in a single 2-D PAGE gel. These polypeptides can be quantified, probed with antibodies (via blotting), tested for posttranslational modifications (using antibodies/chemical stains specific for each PTM), or extracted for mass spectrometric analysis.
After enrichment of various PTMs of interest, they can be profiled via 2-D gel electrophoresis. Some such applications are discussed below.
- Phospho protein expression profiling — phosphorylated protein pIs will shift to a more acidic region on the gel. Phospho expression profiling is used in signal pathway studies
- Acetylated protein expression profiling — acetylation of proteins at their amino termini or on lysine side-chains will cause them to shift to a more acidic region on the gel. Acetylation of proteins can affect protein function, interactions and subsequent or additional posttranslational modifications
- Methylated protein expression profiling — methylation of lysine or arginine residues can have modest effects on protein molecular weight, but shifts their isoelectric points significantly towards the acidic spectrum. Analysis of protein methylation is particularly important in epigenetic studies
- Glycosylated protein expression profiling — glycosylation is both a cotranslational and posttranslational modification. Attachment of relatively simple glycans (O-glycosylation) or more complex glycans (N-glycosylation) can have varying effects on protein molecular weight, pI, and protein functions
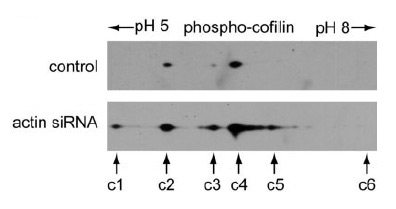
Phospho-cofilin antibody probing of the 2-D gel revealed that spot c6 was the non-phosphorylated cofilin, spots c1–c5 were phosphorylated cofilins. Note that the highly phosphorylated cofilin spot c1 was detectable only in the actin siRNA-treated HeLa cells.


