Description
Description
DCode Systems
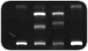
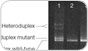
The search for unknown mutations in genomic DNA is important for studying the genetic basis of diseases and disorders, including cancer. Additionally, examining DNA polymorphisms is useful for ecological and evolutionary studies of terrestrial, marine, and microbial organisms, with applications ranging from species identification to delineation of population structure to monitoring genetic diversity.
The DCode universal mutation detection system can scan for single-base changes by any of the following electrophoretic techniques:
- Single-stranded conformation polymorphism (SSCP)
- Denaturing gradient gel electrophoresis (DGGE)
- Constant denaturing gel electrophoresis (CDGE)
- Temporal temperature gradient gel electrophoresis (TTGE)
Flexible and powerful, the DCode system is the one electrophoresis system that can perform any combination of these techniques. At the center of the system is the temperature control module (which includes a microprocessor-controlled heater, a buffer-recirculating pump, and a stirrer). For techniques requiring accurate temperature control, such as SSCP and TTGE, the gels are immersed in the buffer, and temperatures are regulated between 5° and 70°C. Any run temperature below ambient can be achieved with the cooling tank used in conjunction with an external laboratory chiller. The DCode system can run 64 samples in as little as 2 hours — a major consideration when you're screening DNA for sequence variations.
You can configure your DCode system to grow as your needs grow. Each system includes a vertical electrophoresis cell and choice of adaptor kits for SSCP, DGGE, and CDGE, as well as for TTGE, a technique codeveloped by Bio-Rad. TTGE has all the benefits of DGGE and CDGE without chemical denaturant gradients.
This flexibility lets you quickly add new research tools without investing in new instrumentation. Systems configured for the primary application can be expanded to perform any combination of methods by supplementing the core system with technique-specific adaptor kits listed on the accessories tab. Product options include the complete DCode system and individual DCode systems for DDGE, CDGE, TTGE, and SSCP.
WinMelt™ Software
Mutations are most reliably detected when the sequence difference occurs in the lowest melting-temperature domain of the DNA of interest. In addition, optimal resolution is attained only when the molecules do not completely denature. The addition of a GC clamp onto one end of the DNA via PCR ensures that the region screened is in a lower melting-temperature domain, and that the DNA will remain partially double stranded. This enhances both detection and resolution. Windows-based WinMelt software predicts the melting profile of any DNA sequence up to 3,200 bases. Placement of primers and GC clamps can be optimized by analysis of their effect on the DNA melting profile.
Operation of WinMelt software is simple. The DNA sequence is imported from a text file and the melting profile is computed. The data appear onscreen and can be graphed according to user preference. The sequences and melt data can then be exported for use in other programs. WinMelt software is recommended for all DGGE, CDGE, and TTGE applications.
Electrophoresis and Control Reagents
Bio-Rad electrophoresis reagent kits are customized for each application to ensure the highest-quality buffers and acrylamide. Our control reagents for DGGE, CDGE, TTGE, and SSCP help you quickly master any new techniques.
Applications
Technical notes with proven DCode system run conditions are available. To obtain all DCode system application notes, request literature package 1720J. To obtain DCode application notes on microbial diversity studies, request literature package 1720K.
Training Guide
This self-training guide is an interactive CD-ROM that helps you learn about the techniques used to screen mutations using the DCode system. It includes the following:
- Principles of DGGE, CDGE, TTGE, and SSCP
- Videos on setting up and using the DCode system
- WinMelt software tutorial program, DCode application notes, instruction manual, and other literature
To obtain your self-training guide, request catalog #170-9241. The CD-ROM is PC-compatible only.
Note to D GENE™ Users
All of the DCode adaptor kits are compatible with the D GENE apparatus, with the exception of the SSCP adaptor kit. To run gels below room temperature, place the D GENE system in a coldroom or refrigerator equipped with electrical outlets, and use the temperature controller to set the desired run temperature (see bulletin 1935). The basic SSCP adaptor kit is compatible with the D GENE system, but requires use of a coldroom for electrophoresis below room temperature.
Specifications
Specifications
(catalog #170-9080/9083/9086/9089/9092/9095/9098/9102)
Ordering
Ordering
items
Use the filters below to refine results!




Accessories
Accessories















Kits & Reagents
Kits & Reagents